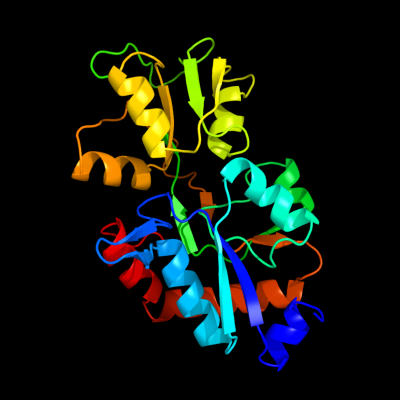
| 1 | c2ylnA_
|
|
 |
100.0 |
35 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter, periplasmic binding protein,
PDBTitle: crystal structure of the l-cystine solute receptor of2 neisseria gonorrhoeae in the closed conformation
|
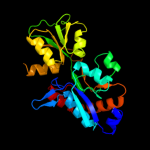
| 2 | c3hv1A_
|
|
 |
100.0 |
25 |
PDB header:transport protein
Chain: A: PDB Molecule:polar amino acid abc uptake transporter substrate
PDBTitle: crystal structure of a polar amino acid abc uptake2 transporter substrate binding protein from streptococcus3 thermophilus
|
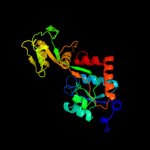
| 3 | c3k4uA_
|
|
 |
100.0 |
31 |
PDB header:transport protein
Chain: A: PDB Molecule:binding component of abc transporter;
PDBTitle: crystal structure of putative binding component of abc transporter2 from wolinella succinogenes dsm 1740 complexed with lysine
|
| 4 | c2ieeB_
|
|
 |
100.0 |
32 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:probable abc transporter extracellular-binding
PDBTitle: crystal structure of yckb_bacsu from bacillus subtilis.2 northeast structural genomics consortium target sr574.
|
| 5 | c3g41A_
|
|
 |
100.0 |
21 |
PDB header:transport protein
Chain: A: PDB Molecule:amino acid abc transporter, periplasmic amino acid-binding
PDBTitle: the structure of cpn0482, the arginine binding protein from the2 periplasm of chlamydia pneumoniae
|
| 6 | c3kbrA_
|
|
 |
100.0 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:cyclohexadienyl dehydratase;
PDBTitle: the crystal structure of cyclohexadienyl dehydratase precursor from2 pseudomonas aeruginosa pa01
|
| 7 | c2o1mB_
|
|
 |
100.0 |
26 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:probable amino-acid abc transporter
PDBTitle: crystal structure of the probable amino-acid abc2 transporter extracellular-binding protein ytmk from3 bacillus subtilis. northeast structural genomics4 consortium target sr572
|
| 8 | c2vhaB_
|
|
 |
100.0 |
22 |
PDB header:transport protein
Chain: B: PDB Molecule:periplasmic binding transport protein;
PDBTitle: debp
|
| 9 | d1hsla_
|
|
 |
100.0 |
27 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 10 | c3r39A_
|
|
 |
100.0 |
24 |
PDB header:transport protein
Chain: A: PDB Molecule:putative periplasmic binding protein;
PDBTitle: crystal structure of periplasmic d-alanine abc transporter from2 salmonella enterica
|
| 11 | c3i6vA_
|
|
 |
100.0 |
28 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic his/glu/gln/arg/opine family-binding protein;
PDBTitle: crystal structure of a periplasmic his/glu/gln/arg/opine family-2 binding protein from silicibacter pomeroyi in complex with lysine
|
| 12 | d1lsta_
|
|
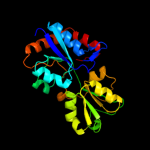 |
100.0 |
27 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 13 | c1xt8B_
|
|
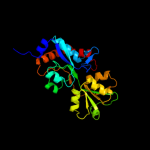 |
100.0 |
21 |
PDB header:transport protein
Chain: B: PDB Molecule:putative amino-acid transporter periplasmic solute-binding
PDBTitle: crystal structure of cysteine-binding protein from campylobacter2 jejuni at 2.0 a resolution
|
| 14 | c3h7mA_
|
|
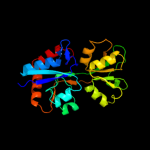 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of a histidine kinase sensor domain with2 similarity to periplasmic binding proteins
|
| 15 | d1wdna_
|
|
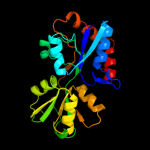 |
100.0 |
32 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
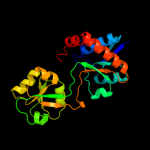
| 16 | c3delC_
|
|
 |
100.0 |
21 |
PDB header:protein binding, transport protein
Chain: C: PDB Molecule:arginine binding protein;
PDBTitle: the structure of ct381, the arginine binding protein from the2 periplasm chlamydia trachomatis
|
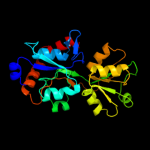
| 17 | c2yjpB_
|
|
 |
100.0 |
28 |
PDB header:transport protein
Chain: B: PDB Molecule:putative abc transporter, periplasmic binding protein,
PDBTitle: crystal structure of the solute receptors for l-cysteine of2 neisseria gonorrhoeae
|
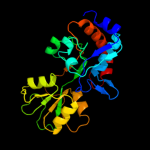
| 18 | c2q89A_
|
|
 |
100.0 |
29 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter amino acid-binding protein;
PDBTitle: crystal structure of ehub in complex with hydroxyectoine
|
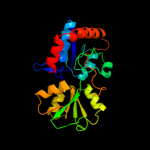
| 19 | c2y7iB_
|
|
 |
100.0 |
29 |
PDB header:arginine-binding protein
Chain: B: PDB Molecule:stm4351;
PDBTitle: structural basis for high arginine specificity in salmonella2 typhimurium periplasmic binding protein stm4351.
|
| 20 | d1xt8a1
|
|
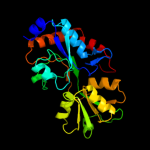 |
100.0 |
22 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 21 | c2q2aD_ |
|
not modelled |
100.0 |
26 |
PDB header:transport protein
Chain: D: PDB Molecule:artj;
PDBTitle: crystal structures of the arginine-, lysine-, histidine-2 binding protein artj from the thermophilic bacterium3 geobacillus stearothermophilus
|
| 22 | c3kzgB_ |
|
not modelled |
100.0 |
27 |
PDB header:transport protein
Chain: B: PDB Molecule:arginine 3rd transport system periplasmic binding
PDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
|
| 23 | c2v25B_ |
|
not modelled |
100.0 |
28 |
PDB header:receptor
Chain: B: PDB Molecule:major cell-binding factor;
PDBTitle: structure of the campylobacter jejuni antigen peb1a, an2 aspartate and glutamate receptor with bound aspartate
|
| 24 | c3mplA_ |
|
not modelled |
100.0 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:virulence sensor protein bvgs;
PDBTitle: crystal structure of bordetella pertussis bvgs vft2 domain (double2 mutant f375e/q461e)
|
| 25 | d1mqia_ |
|
not modelled |
100.0 |
22 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 26 | d2a5sa1 |
|
not modelled |
100.0 |
21 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 27 | c2xx7B_ |
|
not modelled |
100.0 |
22 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamate receptor 2;
PDBTitle: crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-2 (trifluoromethyl)-4,5,6,7-tetrahydro-1h-indazole in complex with3 the ligand binding domain of the rat glua2 receptor and glutamate4 at 2.2a resolution.
|
| 28 | d1ii5a_ |
|
not modelled |
100.0 |
18 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 29 | d1pb7a_ |
|
not modelled |
100.0 |
24 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 30 | d1s50a1 |
|
not modelled |
99.9 |
20 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 31 | c2pyyB_ |
|
not modelled |
99.9 |
21 |
PDB header:transport protein
Chain: B: PDB Molecule:ionotropic glutamate receptor bacterial homologue;
PDBTitle: crystal structure of the glur0 ligand-binding core from nostoc2 punctiforme in complex with (l)-glutamate
|
| 32 | c1yaeB_ |
|
not modelled |
99.9 |
20 |
PDB header:membrane protein
Chain: B: PDB Molecule:glutamate receptor, ionotropic kainate 2;
PDBTitle: structure of the kainate receptor subunit glur6 agonist binding domain2 complexed with domoic acid
|
| 33 | c2rc9A_ |
|
not modelled |
99.9 |
20 |
PDB header:membrane protein
Chain: A: PDB Molecule:glutamate [nmda] receptor subunit 3a;
PDBTitle: crystal structure of the nr3a ligand binding core complex with acpc at2 1.96 angstrom resolution
|
| 34 | c2v3tA_ |
|
not modelled |
99.9 |
20 |
PDB header:receptor
Chain: A: PDB Molecule:glutamate receptor delta-2 subunit synonym
PDBTitle: structure of the ligand-binding core of the ionotropic2 glutamate receptor-like glurdelta2 in the apo form
|
| 35 | d2f34a1 |
|
not modelled |
99.9 |
20 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 36 | c3kg2A_ |
|
not modelled |
99.8 |
27 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:glutamate receptor 2;
PDBTitle: ampa subtype ionotropic glutamate receptor in complex with competitive2 antagonist zk 200775
|
| 37 | c3n5lA_ |
|
not modelled |
98.3 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:binding protein component of abc phosphonate transporter;
PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
|
| 38 | c3e4rA_ |
|
not modelled |
97.7 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein;
PDBTitle: crystal structure of the alkanesulfonate binding protein2 (ssua) from the phytopathogenic bacteria xanthomonas3 axonopodis pv. citri bound to hepes
|
| 39 | d1xs5a_ |
|
not modelled |
97.7 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 40 | c2x26A_ |
|
not modelled |
97.7 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic aliphatic sulphonates-binding protein;
PDBTitle: crystal structure of the periplasmic aliphatic sulphonate2 binding protein ssua from escherichia coli
|
| 41 | c3k2dA_ |
|
not modelled |
97.5 |
13 |
PDB header:immune system
Chain: A: PDB Molecule:abc-type metal ion transport system, periplasmic component;
PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
|
| 42 | c3tqwA_ |
|
not modelled |
97.3 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:methionine-binding protein;
PDBTitle: structure of a abc transporter, periplasmic substrate-binding protein2 from coxiella burnetii
|
| 43 | c3l6gA_ |
|
not modelled |
97.2 |
11 |
PDB header:glycine betaine-binding protein
Chain: A: PDB Molecule:betaine abc transporter permease and substrate binding
PDBTitle: crystal structure of lactococcal opuac in its open conformation
|
| 44 | c2x7pA_ |
|
not modelled |
97.0 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:possible thiamine biosynthesis enzyme;
PDBTitle: the conserved candida albicans ca3427 gene product defines a new2 family of proteins exhibiting the generic periplasmic binding3 protein structural fold
|
| 45 | c3gxaA_ |
|
not modelled |
96.8 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of gna1946
|
| 46 | c3uifA_ |
|
not modelled |
96.8 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:sulfonate abc transporter, periplasmic sulfonate-binding
PDBTitle: crystal structure of putative sulfonate abc transporter, periplasmic2 sulfonate-binding protein ssua from methylobacillus flagellatus kt
|
| 47 | c3ix1B_ |
|
not modelled |
96.5 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding
PDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
|
| 48 | c3ix1A_ |
|
not modelled |
96.5 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding
PDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
|
| 49 | c3ir1F_ |
|
not modelled |
96.5 |
15 |
PDB header:protein binding
Chain: F: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
|
| 50 | c3qslA_ |
|
not modelled |
95.8 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: structure of cae31940 from bordetella bronchiseptica rb50
|
| 51 | c3un6A_ |
|
not modelled |
95.7 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein saouhsc_00137;
PDBTitle: 2.0 angstrom crystal structure of ligand binding component of abc-type2 import system from staphylococcus aureus with zinc bound
|
| 52 | c3tmgA_ |
|
not modelled |
95.1 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine, l-proline abc transporter,
PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
|
| 53 | c2g29A_ |
|
not modelled |
94.7 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein nrta;
PDBTitle: crystal structure of the periplasmic nitrate-binding2 protein nrta from synechocystis pcc 6803
|
| 54 | d1zbma1 |
|
not modelled |
94.5 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 55 | c3ho7A_ |
|
not modelled |
93.8 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:oxyr;
PDBTitle: crystal structure of oxyr from porphyromonas gingivalis
|
| 56 | d1p99a_ |
|
not modelled |
93.8 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 57 | c1p99A_ |
|
not modelled |
93.8 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pg110;
PDBTitle: 1.7a crystal structure of protein pg110 from staphylococcus2 aureus
|
| 58 | c2ql3G_ |
|
not modelled |
92.3 |
11 |
PDB header:transcription
Chain: G: PDB Molecule:probable transcriptional regulator, lysr family protein;
PDBTitle: crystal structure of the c-terminal domain of a probable lysr family2 transcriptional regulator from rhodococcus sp. rha1
|
| 59 | c2i4cA_ |
|
not modelled |
92.1 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:bicarbonate transporter;
PDBTitle: crystal structure of bicarbonate transport protein cmpa from2 synechocystis sp. pcc 6803 in complex with bicarbonate and calcium
|
| 60 | c3n6uA_ |
|
not modelled |
91.6 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:lysr type regulator of tsambcd;
PDBTitle: effector binding domain of tsar in complex with its inducer p-2 toluenesulfonate
|
| 61 | d1i6aa_ |
|
not modelled |
90.4 |
17 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 62 | d1al3a_ |
|
not modelled |
90.4 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 63 | c1al3A_ |
|
not modelled |
90.4 |
12 |
PDB header:transcription regulation
Chain: A: PDB Molecule:cys regulon transcriptional activator cysb;
PDBTitle: cofactor binding fragment of cysb from klebsiella aerogenes
|
| 64 | c3lr1A_ |
|
not modelled |
88.1 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:tungstate abc transporter, periplasmic tungstate-
PDBTitle: the crystal structure of the tungstate abc transporter from2 geobacter sulfurreducens
|
| 65 | c3jv9B_ |
|
not modelled |
88.0 |
12 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, lysr family;
PDBTitle: the structure of a reduced form of oxyr from n. meningitidis
|
| 66 | d2czla1 |
|
not modelled |
83.5 |
11 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 67 | c3muqB_ |
|
not modelled |
82.4 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the crystal structure of a conserved functionally unknown protein from2 vibrio parahaemolyticus rimd 2210633
|
| 68 | c2rejA_ |
|
not modelled |
81.5 |
18 |
PDB header:choline-binding protein
Chain: A: PDB Molecule:putative glycine betaine abc transporter protein;
PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
|
| 69 | c3fd3A_ |
|
not modelled |
78.8 |
7 |
PDB header:transcription regulator
Chain: A: PDB Molecule:chromosome replication initiation inhibitor protein;
PDBTitle: structure of the c-terminal domains of a lysr family protein from2 agrobacterium tumefaciens str. c58.
|
| 70 | c3hn0A_ |
|
not modelled |
77.7 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein;
PDBTitle: crystal structure of an abc transporter (bdi_1369) from2 parabacteroides distasonis at 1.75 a resolution
|
| 71 | c2de4B_ |
|
not modelled |
76.1 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:dibenzothiophene desulfurization enzyme b;
PDBTitle: crystal structure of dszb c27s mutant in complex with biphenyl-2-2 sulfinic acid
|
| 72 | d1amfa_ |
|
not modelled |
74.8 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 73 | c3fj7A_ |
|
not modelled |
68.7 |
16 |
PDB header:protein binding
Chain: A: PDB Molecule:major antigenic peptide peb3;
PDBTitle: crystal structure of l-phospholactate bound peb3
|
| 74 | c3chgB_ |
|
not modelled |
68.4 |
12 |
PDB header:ligand binding protein
Chain: B: PDB Molecule:glycine betaine-binding protein;
PDBTitle: the compatible solute-binding protein opuac from bacillus2 subtilis in complex with dmsa
|
| 75 | d2nxoa1 |
|
not modelled |
63.4 |
10 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 76 | d2esna2 |
|
not modelled |
62.9 |
10 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 77 | c2vd3B_ |
|
not modelled |
60.1 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: the structure of histidine inhibited hisg from2 methanobacterium thermoautotrophicum
|
| 78 | c2uyeA_ |
|
not modelled |
58.0 |
7 |
PDB header:transcription
Chain: A: PDB Molecule:regulatory protein;
PDBTitle: double mutant y110s,f111v dntr from burkholderia sp. strain2 dnt in complex with thiocyanate
|
| 79 | c2h5yC_ |
|
not modelled |
54.1 |
9 |
PDB header:metal transport
Chain: C: PDB Molecule:molybdate-binding periplasmic protein;
PDBTitle: crystallographic structure of the molybdate-binding protein of2 xanthomonas citri at 1.7 ang resolution bound to molybdate
|
| 80 | d1atga_ |
|
not modelled |
54.0 |
11 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 81 | c3b50A_ |
|
not modelled |
53.8 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:sialic acid-binding periplasmic protein siap;
PDBTitle: structure of h. influenzae sialic acid binding protein2 bound to neu5ac.
|
| 82 | c2f78A_ |
|
not modelled |
52.0 |
10 |
PDB header:gene regulation
Chain: A: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: benm effector binding domain with its effector benzoate
|
| 83 | c3hhfB_ |
|
not modelled |
39.9 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, lysr family;
PDBTitle: structure of crga regulatory domain, a lysr-type transcriptional2 regulator from neisseria meningitidis.
|
| 84 | c2h9qC_ |
|
not modelled |
38.3 |
10 |
PDB header:transcription
Chain: C: PDB Molecule:hth-type transcriptional regulator catm;
PDBTitle: crystal structure of the effector binding domain of a catm2 variant (r156h)
|
| 85 | d2ozza1 |
|
not modelled |
38.0 |
9 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 86 | c2dvzA_ |
|
not modelled |
37.9 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: structure of a periplasmic transporter
|
| 87 | d1ixca2 |
|
not modelled |
37.6 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 88 | c2f7cA_ |
|
not modelled |
37.3 |
10 |
PDB header:gene regulation
Chain: A: PDB Molecule:hth-type transcriptional regulator catm;
PDBTitle: catm effector binding domain with its effector cis,cis-muconate
|
| 89 | c2f5xC_ |
|
not modelled |
33.7 |
10 |
PDB header:transport protein
Chain: C: PDB Molecule:bugd;
PDBTitle: structure of periplasmic binding protein bugd
|
| 90 | c3kn3C_ |
|
not modelled |
32.5 |
13 |
PDB header:transcription
Chain: C: PDB Molecule:putative periplasmic protein;
PDBTitle: crystal structure of lysr substrate binding domain (25-263) of2 putative periplasmic protein from wolinella succinogenes
|
| 91 | d1us5a_ |
|
not modelled |
29.3 |
21 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 92 | d2fyia1 |
|
not modelled |
28.9 |
11 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 93 | c3fxbB_ |
|
not modelled |
27.6 |
14 |
PDB header:transport protein
Chain: B: PDB Molecule:trap dicarboxylate transporter, dctp subunit;
PDBTitle: crystal structure of the ectoine-binding protein ueha
|
| 94 | c2hxrA_ |
|
not modelled |
26.6 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional regulator cynr;
PDBTitle: structure of the ligand binding domain of e. coli cynr, a2 transcriptional regulator controlling cyanate metabolism
|
| 95 | c2h9bB_ |
|
not modelled |
26.2 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: crystal structure of the effector binding domain of a benm variant2 (benm r156h/t157s)
|
| 96 | c1jvnB_ |
|
not modelled |
25.8 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 97 | c1twyG_ |
|
not modelled |
24.2 |
12 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:abc transporter, periplasmic substrate-binding protein;
PDBTitle: crystal structure of an abc-type phosphate transport receptor from2 vibrio cholerae
|
| 98 | d1twya_ |
|
not modelled |
24.2 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 99 | d2o38a1 |
|
not modelled |
23.6 |
30 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:NE1354 |
| 100 | c2o38A_ |
|
not modelled |
23.6 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: putative xre family transcriptional regulator
|
| 101 | c3kosA_ |
|
not modelled |
22.2 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional activator ampr;
PDBTitle: structure of the ampr effector binding domain from citrobacter2 freundii
|
| 102 | c2qpqC_ |
|
not modelled |
21.8 |
12 |
PDB header:transport protein
Chain: C: PDB Molecule:protein bug27;
PDBTitle: structure of bug27 from bordetella pertussis
|
| 103 | c3mz1D_ |
|
not modelled |
20.8 |
6 |
PDB header:transcription regulator
Chain: D: PDB Molecule:putative transcriptional regulator;
PDBTitle: the crystal structure of a possible transcription regulator protein2 from sinorhizobium meliloti 1021
|