1 c2pfyA_
100.0
21
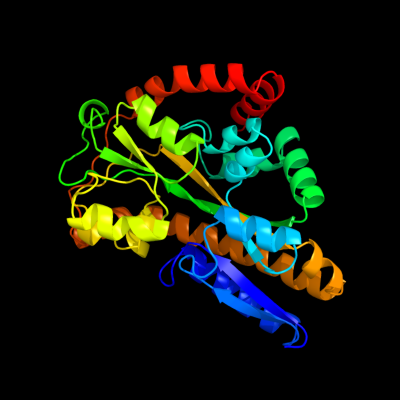
PDB header: transport proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: crystal structure of dctp7, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
2 c3b50A_
100.0
30
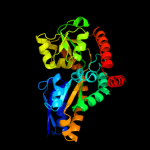
PDB header: transport proteinChain: A: PDB Molecule: sialic acid-binding periplasmic protein siap;PDBTitle: structure of h. influenzae sialic acid binding protein2 bound to neu5ac.
3 c2pfzA_
100.0
22
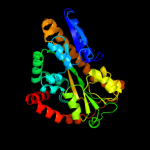
PDB header: transport proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: crystal structure of dctp6, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
4 c3fxbB_
100.0
28
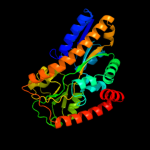
PDB header: transport proteinChain: B: PDB Molecule: trap dicarboxylate transporter, dctp subunit;PDBTitle: crystal structure of the ectoine-binding protein ueha
5 c2vpnB_
100.0
24
PDB header: transportChain: B: PDB Molecule: periplasmic substrate binding protein;PDBTitle: high-resolution structure of the periplasmic ectoine-2 binding protein from teaabc trap-transporter of halomonas3 elongata
6 c2hzkB_
100.0
19
PDB header: ligand binding, transport proteinChain: B: PDB Molecule: trap-t family sorbitol/mannitol transporter, periplasmicPDBTitle: crystal structures of a sodium-alpha-keto acid binding subunit from a2 trap transporter in its open form
7 c2hpgB_
100.0
25
PDB header: ligand binding proteinChain: B: PDB Molecule: abc transporter, periplasmic substrate-bindingPDBTitle: the crystal structure of a thermophilic trap periplasmic2 binding protein
8 c2zzxD_
100.0
20
PDB header: transport proteinChain: D: PDB Molecule: abc transporter, solute-binding protein;PDBTitle: crystal structure of a periplasmic substrate binding protein in2 complex with lactate
9 c3gyyC_
100.0
24
PDB header: transport proteinChain: C: PDB Molecule: periplasmic substrate binding protein;PDBTitle: the ectoine binding protein of the teaabc trap transporter teaa in the2 apo-state
10 c3uifA_
99.5
15
PDB header: transport proteinChain: A: PDB Molecule: sulfonate abc transporter, periplasmic sulfonate-bindingPDBTitle: crystal structure of putative sulfonate abc transporter, periplasmic2 sulfonate-binding protein ssua from methylobacillus flagellatus kt
11 c2x26A_
99.5
18
PDB header: transport proteinChain: A: PDB Molecule: periplasmic aliphatic sulphonates-binding protein;PDBTitle: crystal structure of the periplasmic aliphatic sulphonate2 binding protein ssua from escherichia coli
12 c3e4rA_
99.4
15
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein;PDBTitle: crystal structure of the alkanesulfonate binding protein2 (ssua) from the phytopathogenic bacteria xanthomonas3 axonopodis pv. citri bound to hepes
13 c2x7pA_
99.2
10
PDB header: unknown functionChain: A: PDB Molecule: possible thiamine biosynthesis enzyme;PDBTitle: the conserved candida albicans ca3427 gene product defines a new2 family of proteins exhibiting the generic periplasmic binding3 protein structural fold
14 c3qslA_
99.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative exported protein;PDBTitle: structure of cae31940 from bordetella bronchiseptica rb50
15 c3ix1B_
99.2
14
PDB header: biosynthetic proteinChain: B: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
16 c3ix1A_
99.2
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
17 c2de4B_
99.1
16
PDB header: hydrolaseChain: B: PDB Molecule: dibenzothiophene desulfurization enzyme b;PDBTitle: crystal structure of dszb c27s mutant in complex with biphenyl-2-2 sulfinic acid
18 c3un6A_
99.1
16
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein saouhsc_00137;PDBTitle: 2.0 angstrom crystal structure of ligand binding component of abc-type2 import system from staphylococcus aureus with zinc bound
19 c3hn0A_
99.0
9
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein;PDBTitle: crystal structure of an abc transporter (bdi_1369) from2 parabacteroides distasonis at 1.75 a resolution
20 d1us5a_
99.0
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like21 d1p99a_
not modelled
98.9
17
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like22 c1p99A_
not modelled
98.9
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pg110;PDBTitle: 1.7a crystal structure of protein pg110 from staphylococcus2 aureus
23 c3n5lA_
not modelled
98.8
15
PDB header: transport proteinChain: A: PDB Molecule: binding protein component of abc phosphonate transporter;PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
24 c3gxaA_
not modelled
98.8
16
PDB header: protein bindingChain: A: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of gna1946
25 c2g29A_
not modelled
98.8
14
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein nrta;PDBTitle: crystal structure of the periplasmic nitrate-binding2 protein nrta from synechocystis pcc 6803
26 c3k2dA_
not modelled
98.8
16
PDB header: immune systemChain: A: PDB Molecule: abc-type metal ion transport system, periplasmic component;PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
27 c3tmgA_
not modelled
98.8
13
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine, l-proline abc transporter,PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
28 d1xs5a_
not modelled
98.8
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like29 d2czla1
not modelled
98.7
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like30 c2i4cA_
not modelled
98.7
17
PDB header: transport proteinChain: A: PDB Molecule: bicarbonate transporter;PDBTitle: crystal structure of bicarbonate transport protein cmpa from2 synechocystis sp. pcc 6803 in complex with bicarbonate and calcium
31 d1zbma1
not modelled
98.6
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like32 c2qpqC_
not modelled
98.6
9
PDB header: transport proteinChain: C: PDB Molecule: protein bug27;PDBTitle: structure of bug27 from bordetella pertussis
33 c3tqwA_
not modelled
98.5
19
PDB header: transport proteinChain: A: PDB Molecule: methionine-binding protein;PDBTitle: structure of a abc transporter, periplasmic substrate-binding protein2 from coxiella burnetii
34 c3l6gA_
not modelled
98.5
14
PDB header: glycine betaine-binding proteinChain: A: PDB Molecule: betaine abc transporter permease and substrate bindingPDBTitle: crystal structure of lactococcal opuac in its open conformation
35 d2nxoa1
not modelled
98.4
12
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like36 c3ir1F_
not modelled
98.3
17
PDB header: protein bindingChain: F: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
37 c2dvzA_
not modelled
98.2
14
PDB header: transport proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: structure of a periplasmic transporter
38 c2f5xC_
not modelled
98.2
11
PDB header: transport proteinChain: C: PDB Molecule: bugd;PDBTitle: structure of periplasmic binding protein bugd
39 c1q1kA_
not modelled
97.1
16
PDB header: transferaseChain: A: PDB Molecule: atp phosphoribosyltransferase;PDBTitle: structure of atp-phosphoribosyltransferase from e. coli complexed with2 pr-atp
40 c3r6uA_
not modelled
97.1
16
PDB header: transport proteinChain: A: PDB Molecule: choline-binding protein;PDBTitle: crystal structure of choline binding protein opubc from bacillus2 subtilis
41 d2a5sa1
not modelled
97.1
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like42 c2rejA_
not modelled
96.9
14
PDB header: choline-binding proteinChain: A: PDB Molecule: putative glycine betaine abc transporter protein;PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
43 d1h3da1
not modelled
96.9
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like44 c3kzgB_
not modelled
96.8
11
PDB header: transport proteinChain: B: PDB Molecule: arginine 3rd transport system periplasmic bindingPDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
45 c3pppA_
not modelled
96.8
14
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine/carnitine/choline-binding protein;PDBTitle: structures of the substrate-binding protein provide insights into the2 multiple compatible solutes binding specificities of bacillus3 subtilis abc transporter opuc
46 c2rc9A_
not modelled
96.5
13
PDB header: membrane proteinChain: A: PDB Molecule: glutamate [nmda] receptor subunit 3a;PDBTitle: crystal structure of the nr3a ligand binding core complex with acpc at2 1.96 angstrom resolution
47 c2vd3B_
not modelled
96.5
13
PDB header: transferaseChain: B: PDB Molecule: atp phosphoribosyltransferase;PDBTitle: the structure of histidine inhibited hisg from2 methanobacterium thermoautotrophicum
48 c2o1mB_
not modelled
96.4
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable amino-acid abc transporterPDBTitle: crystal structure of the probable amino-acid abc2 transporter extracellular-binding protein ytmk from3 bacillus subtilis. northeast structural genomics4 consortium target sr572
49 c3o66A_
not modelled
96.4
12
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine/carnitine/choline abc transporter;PDBTitle: crystal structure of glycine betaine/carnitine/choline abc transporter
50 c2ylnA_
not modelled
96.3
19
PDB header: transport proteinChain: A: PDB Molecule: putative abc transporter, periplasmic binding protein,PDBTitle: crystal structure of the l-cystine solute receptor of2 neisseria gonorrhoeae in the closed conformation
51 c1nh7A_
not modelled
96.3
18
PDB header: transferaseChain: A: PDB Molecule: atp phosphoribosyltransferase;PDBTitle: atp phosphoribosyltransferase (atp-prtase) from mycobacterium2 tuberculosis
52 d1pb7a_
not modelled
96.1
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like53 d1sw5a_
not modelled
95.2
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like54 c2q2aD_
not modelled
95.1
15
PDB header: transport proteinChain: D: PDB Molecule: artj;PDBTitle: crystal structures of the arginine-, lysine-, histidine-2 binding protein artj from the thermophilic bacterium3 geobacillus stearothermophilus
55 d1z7me1
not modelled
94.1
17
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like56 d1nh8a1
not modelled
93.7
17
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like57 c2pyyB_
not modelled
92.8
13
PDB header: transport proteinChain: B: PDB Molecule: ionotropic glutamate receptor bacterial homologue;PDBTitle: crystal structure of the glur0 ligand-binding core from nostoc2 punctiforme in complex with (l)-glutamate
58 c3k4uA_
not modelled
91.9
22
PDB header: transport proteinChain: A: PDB Molecule: binding component of abc transporter;PDBTitle: crystal structure of putative binding component of abc transporter2 from wolinella succinogenes dsm 1740 complexed with lysine
59 d1vmda_
not modelled
90.6
21
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA60 c2ypnA_
not modelled
89.8
16
PDB header: transferaseChain: A: PDB Molecule: protein (hydroxymethylbilane synthase);PDBTitle: hydroxymethylbilane synthase
61 c3mplA_
not modelled
89.5
16
PDB header: signaling proteinChain: A: PDB Molecule: virulence sensor protein bvgs;PDBTitle: crystal structure of bordetella pertussis bvgs vft2 domain (double2 mutant f375e/q461e)
62 d1lsta_
not modelled
89.2
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like63 d1ii5a_
not modelled
88.7
11
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like64 c2y7iB_
not modelled
88.6
16
PDB header: arginine-binding proteinChain: B: PDB Molecule: stm4351;PDBTitle: structural basis for high arginine specificity in salmonella2 typhimurium periplasmic binding protein stm4351.
65 d2ozza1
not modelled
87.2
10
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like66 d1pdaa1
not modelled
86.5
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like67 c3r39A_
not modelled
85.6
12
PDB header: transport proteinChain: A: PDB Molecule: putative periplasmic binding protein;PDBTitle: crystal structure of periplasmic d-alanine abc transporter from2 salmonella enterica
68 d1ve4a1
not modelled
85.1
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like69 d1r9la_
not modelled
84.5
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like70 c2vd2A_
not modelled
83.3
10
PDB header: transferaseChain: A: PDB Molecule: atp phosphoribosyltransferase;PDBTitle: the crystal structure of hisg from b. subtilis
71 c3eq1A_
not modelled
82.7
14
PDB header: transferaseChain: A: PDB Molecule: porphobilinogen deaminase;PDBTitle: the crystal structure of human porphobilinogen deaminase at2 2.8a resolution
72 d1b1xa1
not modelled
82.3
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin73 c3lr1A_
not modelled
81.9
17
PDB header: transport proteinChain: A: PDB Molecule: tungstate abc transporter, periplasmic tungstate-PDBTitle: the crystal structure of the tungstate abc transporter from2 geobacter sulfurreducens
74 c3g41A_
not modelled
80.9
14
PDB header: transport proteinChain: A: PDB Molecule: amino acid abc transporter, periplasmic amino acid-bindingPDBTitle: the structure of cpn0482, the arginine binding protein from the2 periplasm of chlamydia pneumoniae
75 c3kn3C_
not modelled
80.7
19
PDB header: transcriptionChain: C: PDB Molecule: putative periplasmic protein;PDBTitle: crystal structure of lysr substrate binding domain (25-263) of2 putative periplasmic protein from wolinella succinogenes
76 c1y4cA_
not modelled
79.9
14
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
77 d1b93a_
not modelled
78.8
15
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA78 d1eh3a_
not modelled
78.1
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin79 d1o63a_
not modelled
78.0
11
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like80 d1hsla_
not modelled
77.0
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like81 c2v25B_
not modelled
76.6
15
PDB header: receptorChain: B: PDB Molecule: major cell-binding factor;PDBTitle: structure of the campylobacter jejuni antigen peb1a, an2 aspartate and glutamate receptor with bound aspartate
82 d1ryoa_
not modelled
76.2
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin83 c3luyA_
not modelled
73.5
14
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
84 d1dtza1
not modelled
72.8
17
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin85 d1sbpa_
not modelled
71.8
9
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like86 c3delC_
not modelled
70.7
13
PDB header: protein binding, transport proteinChain: C: PDB Molecule: arginine binding protein;PDBTitle: the structure of ct381, the arginine binding protein from the2 periplasm chlamydia trachomatis
87 d2i6ea1
not modelled
69.0
11
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like88 d1wo8a1
not modelled
68.5
16
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA89 c3hv1A_
not modelled
65.9
15
PDB header: transport proteinChain: A: PDB Molecule: polar amino acid abc uptake transporter substratePDBTitle: crystal structure of a polar amino acid abc uptake2 transporter substrate binding protein from streptococcus3 thermophilus
90 c2z8fB_
not modelled
63.2
9
PDB header: sugar binding proteinChain: B: PDB Molecule: galacto-n-biose/lacto-n-biose i transporter substrate-PDBTitle: the galacto-n-biose-/lacto-n-biose i-binding protein (gl-bp) of the2 abc transporter from bifidobacterium longum in complex with lacto-n-3 tetraose
91 c2i6eG_
not modelled
58.3
12
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of protein dr0370 from deinococcus radiodurans, pfam2 duf178
92 d2qmwa1
not modelled
56.8
12
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like93 c3h7mA_
not modelled
56.2
12
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of a histidine kinase sensor domain with2 similarity to periplasmic binding proteins
94 c3c4mA_
not modelled
55.6
16
PDB header: membrane proteinChain: A: PDB Molecule: fusion protein of maltose-binding periplasmic protein andPDBTitle: structure of human parathyroid hormone in complex with the2 extracellular domain of its g-protein-coupled receptor (pth1r)
95 c3kbrA_
not modelled
55.0
10
PDB header: lyaseChain: A: PDB Molecule: cyclohexadienyl dehydratase;PDBTitle: the crystal structure of cyclohexadienyl dehydratase precursor from2 pseudomonas aeruginosa pa01
96 c3ob4A_
not modelled
54.8
14
PDB header: allergenChain: A: PDB Molecule: maltose abc transporter periplasmic protein, arah 2;PDBTitle: mbp-fusion protein of the major peanut allergen ara h 2
97 d1tfda_
not modelled
54.0
20
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin98 c1hsjA_
not modelled
53.8
13
PDB header: transcription/sugar binding proteinChain: A: PDB Molecule: fusion protein consisting of staphylococcusPDBTitle: sarr mbp fusion structure
99 d1y9ba1
not modelled
53.8
16
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: VCA0319-like100 d2hava1
not modelled
53.3
22
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin101 c3oaiB_
not modelled
51.0
14
PDB header: membrane protein, cell adhesionChain: B: PDB Molecule: maltose-binding periplasmic protein, myelin protein p0;PDBTitle: crystal structure of the extra-cellular domain of human myelin protein2 zero
102 c1r6zA_
not modelled
48.9
14
PDB header: gene regulationChain: A: PDB Molecule: chimera of maltose-binding periplasmic protein andPDBTitle: the crystal structure of the argonaute2 paz domain (as a mbp fusion)
103 c2h9bB_
not modelled
48.5
15
PDB header: transcriptionChain: B: PDB Molecule: hth-type transcriptional regulator benm;PDBTitle: crystal structure of the effector binding domain of a benm variant2 (benm r156h/t157s)
104 c3osrA_
not modelled
48.3
15
PDB header: fluorescent protein, transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, green fluorescentPDBTitle: maltose-bound maltose sensor engineered by insertion of circularly2 permuted green fluorescent protein into e. coli maltose binding3 protein at position 311
105 c3muqB_
not modelled
47.8
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized conserved protein;PDBTitle: the crystal structure of a conserved functionally unknown protein from2 vibrio parahaemolyticus rimd 2210633
106 c3d4cA_
not modelled
46.6
14
PDB header: cell adhesionChain: A: PDB Molecule: maltose-binding periplasmic protein, linker, zona pellucidaPDBTitle: zp-n domain of mammalian sperm receptor zp3 (crystal form i)
107 d1ieja_
not modelled
45.8
22
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin108 c3f5fA_
not modelled
45.2
13
PDB header: transport, transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein, heparanPDBTitle: crystal structure of heparan sulfate 2-o-sulfotransferase2 from gallus gallus as a maltose binding protein fusion.
109 d1jnfa1
not modelled
44.8
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin110 d1wdna_
not modelled
44.0
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like111 d1h76a1
not modelled
42.3
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin112 c2qmwA_
not modelled
42.2
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of the prephenate dehydratase (pdt) from2 staphylococcus aureus subsp. aureus mu50
113 c2ieeB_
not modelled
42.1
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable abc transporter extracellular-bindingPDBTitle: crystal structure of yckb_bacsu from bacillus subtilis.2 northeast structural genomics consortium target sr574.
114 c3i6vA_
not modelled
41.7
8
PDB header: transport proteinChain: A: PDB Molecule: periplasmic his/glu/gln/arg/opine family-binding protein;PDBTitle: crystal structure of a periplasmic his/glu/gln/arg/opine family-2 binding protein from silicibacter pomeroyi in complex with lysine
115 c2q89A_
not modelled
40.8
13
PDB header: transport proteinChain: A: PDB Molecule: putative abc transporter amino acid-binding protein;PDBTitle: crystal structure of ehub in complex with hydroxyectoine
116 d2iiza1
not modelled
39.5
14
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Dyp-type peroxidase-like117 c3fj7A_
not modelled
39.4
13
PDB header: protein bindingChain: A: PDB Molecule: major antigenic peptide peb3;PDBTitle: crystal structure of l-phospholactate bound peb3
118 c3fd3A_
not modelled
38.0
14
PDB header: transcription regulatorChain: A: PDB Molecule: chromosome replication initiation inhibitor protein;PDBTitle: structure of the c-terminal domains of a lysr family protein from2 agrobacterium tumefaciens str. c58.
119 c2vgqA_
not modelled
37.8
14
PDB header: immune system/transportChain: A: PDB Molecule: maltose-binding periplasmic protein,PDBTitle: crystal structure of human ips-1 card
120 d1pc3a_
not modelled
36.9
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like