1 c2o7jA_
100.0
19
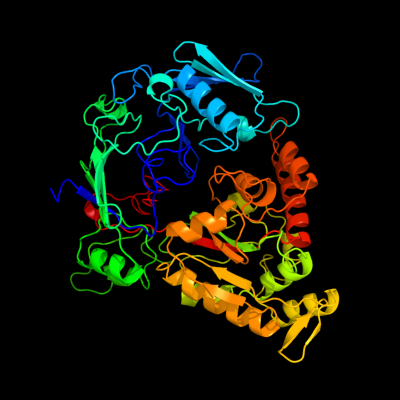
PDB header: sugar binding proteinChain: A: PDB Molecule: oligopeptide abc transporter, periplasmicPDBTitle: the x-ray crystal structure of a thermophilic cellobiose2 binding protein bound with cellopentaose
2 c1ztyA_
100.0
18
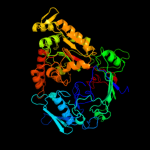
PDB header: sugar binding protein, signaling proteinChain: A: PDB Molecule: chitin oligosaccharide binding protein;PDBTitle: crystal structure of the chitin oligasaccharide binding2 protein
3 d1xoca1
100.0
21
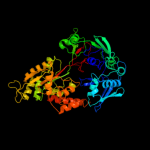
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like4 c2wokA_
100.0
20
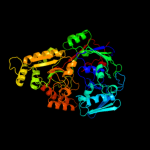
PDB header: peptide binding protein/peptideChain: A: PDB Molecule: clavulanic acid biosynthesis oligopeptidePDBTitle: clavulanic acid biosynthesis oligopeptide2 binding protein 2 complexed with bradykinin
5 d1vr5a1
100.0
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like6 d1zlqa1
100.0
20
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like7 d1dpea_
100.0
17
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like8 c3tpaA_
100.0
18
PDB header: heme binding proteinChain: A: PDB Molecule: heme-binding protein a;PDBTitle: structure of hbpa2 from haemophilus parasuis
9 c3m8uA_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: heme-binding protein a;PDBTitle: crystal structure of glutathione-binding protein a (gbpa) from2 haemophilus parasuis sh0165 in complex with glutathione disulfide3 (gssg)
10 d1jeta_
100.0
17
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like11 c3t66A_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: nickel abc transporter (nickel-binding protein);PDBTitle: crystal structure of nickel abc transporter from bacillus halodurans
12 c3o9pA_
100.0
14
PDB header: peptide binding protein/peptideChain: A: PDB Molecule: periplasmic murein peptide-binding protein;PDBTitle: the structure of the escherichia coli murein tripeptide binding2 protein mppa
13 d1uqwa_
100.0
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like14 c2grvC_
100.0
16
PDB header: biosynthetic proteinChain: C: PDB Molecule: lpqw;PDBTitle: crystal structure of lpqw
15 c3ftoA_
100.0
19
PDB header: peptide binding proteinChain: A: PDB Molecule: oligopeptide-binding protein oppa;PDBTitle: crystal structure of oppa in a open conformation
16 c3ry3B_
100.0
20
PDB header: transport proteinChain: B: PDB Molecule: putative solute-binding protein;PDBTitle: putative solute-binding protein from yersinia pestis.
17 c2d5wA_
100.0
17
PDB header: peptide binding proteinChain: A: PDB Molecule: peptide abc transporter, peptide-binding protein;PDBTitle: the crystal structure of oligopeptide binding protein from thermus2 thermophilus hb8 complexed with pentapeptide
18 c3rqtA_
100.0
17
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: 1.5 angstrom crystal structure of the complex of ligand binding2 component of abc-type import system from staphylococcus aureus with3 nickel and two histidines
19 c3lvuB_
100.0
35
PDB header: transport proteinChain: B: PDB Molecule: abc transporter, periplasmic substrate-binding protein;PDBTitle: crystal structure of abc transporter, periplasmic substrate-binding2 protein spo2066 from silicibacter pomeroyi
20 c3pamB_
100.0
35
PDB header: transport proteinChain: B: PDB Molecule: transmembrane protein;PDBTitle: crystal structure of a domain of transmembrane protein of abc-type2 oligopeptide transport system from bartonella henselae str. houston-1
21 c3o6pA_
not modelled
100.0
19
PDB header: protein bindingChain: A: PDB Molecule: peptide abc transporter, peptide-binding protein;PDBTitle: crystal structure of peptide abc transporter, peptide-binding protein
22 c3chgB_
not modelled
96.3
15
PDB header: ligand binding proteinChain: B: PDB Molecule: glycine betaine-binding protein;PDBTitle: the compatible solute-binding protein opuac from bacillus2 subtilis in complex with dmsa
23 c3l6gA_
not modelled
95.8
12
PDB header: glycine betaine-binding proteinChain: A: PDB Molecule: betaine abc transporter permease and substrate bindingPDBTitle: crystal structure of lactococcal opuac in its open conformation
24 c3tmgA_
not modelled
95.6
13
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine, l-proline abc transporter,PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
25 d1r9la_
not modelled
93.7
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like26 c2rejA_
not modelled
93.2
12
PDB header: choline-binding proteinChain: A: PDB Molecule: putative glycine betaine abc transporter protein;PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
27 d2p0la1
not modelled
92.6
17
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like28 c3nohA_
not modelled
92.6
17
PDB header: peptide binding proteinChain: A: PDB Molecule: putative peptide binding protein;PDBTitle: crystal structure of a putative peptide binding protein (rumgna_00914)2 from ruminococcus gnavus atcc 29149 at 1.60 a resolution
29 c3r6uA_
not modelled
87.8
9
PDB header: transport proteinChain: A: PDB Molecule: choline-binding protein;PDBTitle: crystal structure of choline binding protein opubc from bacillus2 subtilis
30 d2fz5a1
not modelled
82.1
3
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related31 c3k2dA_
not modelled
82.0
7
PDB header: immune systemChain: A: PDB Molecule: abc-type metal ion transport system, periplasmic component;PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
32 d1sw5a_
not modelled
81.5
7
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like33 c3ir1F_
not modelled
80.7
11
PDB header: protein bindingChain: F: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
34 d1f4pa_
not modelled
79.6
9
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related35 d2hfqa1
not modelled
77.9
15
Fold: NE1680-likeSuperfamily: NE1680-likeFamily: NE1680-like36 c2hfqA_
not modelled
77.9
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: nmr structure of protein ne1680 from nitrosomonas europaea:2 northeast structural genomics consortium target net5
37 c3gxaA_
not modelled
77.7
11
PDB header: protein bindingChain: A: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of gna1946
38 d1ykga1
not modelled
77.3
9
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like39 c2uzhB_
not modelled
77.0
16
PDB header: lyaseChain: B: PDB Molecule: 2c-methyl-d-erythritol 2,4-cyclodiphosphatePDBTitle: mycobacterium smegmatis 2c-methyl-d-erythritol-2,4-2 cyclodiphosphate synthase (ispf)
40 d1b1ca_
not modelled
75.9
5
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like41 d5nula_
not modelled
74.0
6
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related42 d1xs5a_
not modelled
73.1
7
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like43 c3hlyA_
not modelled
71.3
4
PDB header: flavoproteinChain: A: PDB Molecule: flavodoxin-like domain;PDBTitle: crystal structure of the flavodoxin-like domain from2 synechococcus sp q5mzp6_synp6 protein. northeast structural3 genomics consortium target snr135d.
44 c3kzgB_
not modelled
70.1
11
PDB header: transport proteinChain: B: PDB Molecule: arginine 3rd transport system periplasmic bindingPDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
45 d1ja1a2
not modelled
69.5
5
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like46 c3f6sI_
not modelled
69.4
7
PDB header: electron transportChain: I: PDB Molecule: flavodoxin;PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin2 alternate conformers
47 c3ombA_
not modelled
69.2
13
PDB header: transport proteinChain: A: PDB Molecule: extracellular solute-binding protein, family 1;PDBTitle: crystal structure of extracellular solute-binding protein from2 bifidobacterium longum subsp. infantis
48 c2bpoA_
not modelled
68.8
8
PDB header: reductaseChain: A: PDB Molecule: nadph-cytochrom p450 reductase;PDBTitle: crystal structure of the yeast cpr triple mutant: d74g,2 y75f, k78a.
49 d1ycga1
not modelled
64.8
8
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related50 c3fniA_
not modelled
63.9
5
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
51 c3n5lA_
not modelled
62.5
17
PDB header: transport proteinChain: A: PDB Molecule: binding protein component of abc phosphonate transporter;PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
52 d1bvyf_
not modelled
61.7
9
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related53 c1bvyF_
not modelled
61.7
9
PDB header: oxidoreductaseChain: F: PDB Molecule: protein (cytochrome p450 bm-3);PDBTitle: complex of the heme and fmn-binding domains of the2 cytochrome p450(bm-3)
54 c2rc9A_
not modelled
61.6
11
PDB header: membrane proteinChain: A: PDB Molecule: glutamate [nmda] receptor subunit 3a;PDBTitle: crystal structure of the nr3a ligand binding core complex with acpc at2 1.96 angstrom resolution
55 c3pppA_
not modelled
61.3
11
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine/carnitine/choline-binding protein;PDBTitle: structures of the substrate-binding protein provide insights into the2 multiple compatible solutes binding specificities of bacillus3 subtilis abc transporter opuc
56 c2p3nB_
not modelled
60.5
17
PDB header: hydrolaseChain: B: PDB Molecule: inositol-1-monophosphatase;PDBTitle: thermotoga maritima impase tm1415
57 d1iv3a_
not modelled
60.0
27
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like58 c3kn3C_
not modelled
58.3
9
PDB header: transcriptionChain: C: PDB Molecule: putative periplasmic protein;PDBTitle: crystal structure of lysr substrate binding domain (25-263) of2 putative periplasmic protein from wolinella succinogenes
59 c2pmpA_
not modelled
56.6
25
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from2 the isoprenoid biosynthetic pathway of arabidopsis thaliana
60 c1j9zB_
not modelled
56.4
7
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-cytochrome p450 reductase;PDBTitle: cypor-w677g
61 c3f0gA_
not modelled
56.3
12
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: co-crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate2 synthase with cmp
62 d1e5da1
not modelled
56.2
7
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related63 d1vh8a_
not modelled
55.9
14
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like64 d1w55a2
not modelled
55.4
16
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like65 d1t0aa_
not modelled
54.5
17
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like66 c3hr4C_
not modelled
53.6
10
PDB header: oxidoreductase/metal binding proteinChain: C: PDB Molecule: nitric oxide synthase, inducible;PDBTitle: human inos reductase and calmodulin complex
67 c3lr1A_
not modelled
50.3
11
PDB header: transport proteinChain: A: PDB Molecule: tungstate abc transporter, periplasmic tungstate-PDBTitle: the crystal structure of the tungstate abc transporter from2 geobacter sulfurreducens
68 d1p99a_
not modelled
50.1
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like69 c1p99A_
not modelled
50.1
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pg110;PDBTitle: 1.7a crystal structure of protein pg110 from staphylococcus2 aureus
70 d1gx1a_
not modelled
49.8
16
Fold: Bacillus chorismate mutase-likeSuperfamily: IpsF-likeFamily: IpsF-like71 d1vmea1
not modelled
49.6
8
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related72 c3re3B_
not modelled
49.0
18
PDB header: lyaseChain: B: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: crystal structure of 2-c-methyl-d-erythritol 2,4-cyclodiphosphate2 synthase from francisella tularensis
73 d1xvya_
not modelled
48.1
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like74 c3nbmA_
not modelled
47.8
13
PDB header: transferaseChain: A: PDB Molecule: pts system, lactose-specific iibc components;PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
75 c3s83A_
not modelled
47.7
13
PDB header: signaling proteinChain: A: PDB Molecule: ggdef family protein;PDBTitle: crystal structure of eal domain from caulobacter crescentus cb15
76 d2a5sa1
not modelled
47.5
7
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like77 d1twya_
not modelled
47.2
6
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like78 c1twyG_
not modelled
47.2
6
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: abc transporter, periplasmic substrate-binding protein;PDBTitle: crystal structure of an abc-type phosphate transport receptor from2 vibrio cholerae
79 d1tlla2
not modelled
46.0
3
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like80 c2y7iB_
not modelled
43.1
5
PDB header: arginine-binding proteinChain: B: PDB Molecule: stm4351;PDBTitle: structural basis for high arginine specificity in salmonella2 typhimurium periplasmic binding protein stm4351.
81 c3r39A_
not modelled
43.1
11
PDB header: transport proteinChain: A: PDB Molecule: putative periplasmic binding protein;PDBTitle: crystal structure of periplasmic d-alanine abc transporter from2 salmonella enterica
82 d1nnfa_
not modelled
42.8
8
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like83 c3muqB_
not modelled
42.0
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized conserved protein;PDBTitle: the crystal structure of a conserved functionally unknown protein from2 vibrio parahaemolyticus rimd 2210633
84 c2hnbA_
not modelled
41.8
5
PDB header: electron transportChain: A: PDB Molecule: protein mioc;PDBTitle: solution structure of a bacterial holo-flavodoxin
85 d1xc1a_
not modelled
41.6
12
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like86 c2axoA_
not modelled
41.0
18
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein atu2684;PDBTitle: x-ray crystal structure of protein agr_c_4864 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr35.
87 c2ylnA_
not modelled
39.7
9
PDB header: transport proteinChain: A: PDB Molecule: putative abc transporter, periplasmic binding protein,PDBTitle: crystal structure of the l-cystine solute receptor of2 neisseria gonorrhoeae in the closed conformation
88 c2f5xC_
not modelled
39.5
10
PDB header: transport proteinChain: C: PDB Molecule: bugd;PDBTitle: structure of periplasmic binding protein bugd
89 d1wdna_
not modelled
39.4
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like90 c2wc1A_
not modelled
39.0
5
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
91 c3g41A_
not modelled
37.7
9
PDB header: transport proteinChain: A: PDB Molecule: amino acid abc transporter, periplasmic amino acid-bindingPDBTitle: the structure of cpn0482, the arginine binding protein from the2 periplasm of chlamydia pneumoniae
92 c3hn0A_
not modelled
36.5
7
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein;PDBTitle: crystal structure of an abc transporter (bdi_1369) from2 parabacteroides distasonis at 1.75 a resolution
93 c2q2aD_
not modelled
36.2
11
PDB header: transport proteinChain: D: PDB Molecule: artj;PDBTitle: crystal structures of the arginine-, lysine-, histidine-2 binding protein artj from the thermophilic bacterium3 geobacillus stearothermophilus
94 c3k4uA_
not modelled
36.2
7
PDB header: transport proteinChain: A: PDB Molecule: binding component of abc transporter;PDBTitle: crystal structure of putative binding component of abc transporter2 from wolinella succinogenes dsm 1740 complexed with lysine
95 d2f06a2
not modelled
36.0
27
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like96 c3b6nA_
not modelled
35.2
17
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphatePDBTitle: crystal structure of 2c-methyl-d-erythritol 2,4-2 cyclodiphosphate synthase pv003920 from plasmodium vivax
97 c2q9uB_
not modelled
34.6
11
PDB header: oxidoreductaseChain: B: PDB Molecule: a-type flavoprotein;PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
98 d1hsla_
not modelled
33.5
5
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like99 c1vmeB_
not modelled
33.5
10
PDB header: electron transportChain: B: PDB Molecule: flavoprotein;PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
100 c2x5dD_
not modelled
33.0
18
PDB header: transferaseChain: D: PDB Molecule: probable aminotransferase;PDBTitle: crystal structure of a probable aminotransferase from2 pseudomonas aeruginosa
101 c3tqwA_
not modelled
32.5
13
PDB header: transport proteinChain: A: PDB Molecule: methionine-binding protein;PDBTitle: structure of a abc transporter, periplasmic substrate-binding protein2 from coxiella burnetii
102 c2xx7B_
not modelled
32.5
17
PDB header: transport proteinChain: B: PDB Molecule: glutamate receptor 2;PDBTitle: crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-2 (trifluoromethyl)-4,5,6,7-tetrahydro-1h-indazole in complex with3 the ligand binding domain of the rat glua2 receptor and glutamate4 at 2.2a resolution.
103 c1w57A_
not modelled
32.2
15
PDB header: transferaseChain: A: PDB Molecule: ispd/ispf bifunctional enzyme;PDBTitle: structure of the bifunctional ispdf from campylobacter2 jejuni containing zn
104 c3luuA_
not modelled
31.4
15
PDB header: biosynthetic proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with unknown function which belongs to2 pfam duf971 family (afe_2189) from acidithiobacillus ferrooxidans3 atcc 23270 at 1.93 a resolution
105 d1xvxa_
not modelled
31.1
13
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like106 c3delC_
not modelled
31.1
5
PDB header: protein binding, transport proteinChain: C: PDB Molecule: arginine binding protein;PDBTitle: the structure of ct381, the arginine binding protein from the2 periplasm chlamydia trachomatis
107 d2axoa1
not modelled
30.8
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Atu2684-like108 d1pb7a_
not modelled
30.4
11
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like109 c2l2qA_
not modelled
29.4
16
PDB header: transferaseChain: A: PDB Molecule: pts system, cellobiose-specific iib component (cela);PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
110 c2ek8A_
not modelled
29.1
6
PDB header: hydrolaseChain: A: PDB Molecule: aminopeptidase;PDBTitle: aminopeptidase from aneurinibacillus sp. strain am-1
111 d1us5a_
not modelled
27.9
9
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like112 c2gquA_
not modelled
27.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-n-acetylenolpyruvylglucosamine reductase;PDBTitle: crystal structure of udp-n-acetylenolpyruvylglucosamine2 reductase (murb) from thermus caldophilus
113 c3i6vA_
not modelled
26.8
18
PDB header: transport proteinChain: A: PDB Molecule: periplasmic his/glu/gln/arg/opine family-binding protein;PDBTitle: crystal structure of a periplasmic his/glu/gln/arg/opine family-2 binding protein from silicibacter pomeroyi in complex with lysine
114 c2qpqC_
not modelled
26.1
12
PDB header: transport proteinChain: C: PDB Molecule: protein bug27;PDBTitle: structure of bug27 from bordetella pertussis
115 d1lsta_
not modelled
25.7
7
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like116 d1h3da1
not modelled
25.6
13
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like117 c2ieeB_
not modelled
24.2
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable abc transporter extracellular-bindingPDBTitle: crystal structure of yckb_bacsu from bacillus subtilis.2 northeast structural genomics consortium target sr574.
118 c1q1kA_
not modelled
23.7
13
PDB header: transferaseChain: A: PDB Molecule: atp phosphoribosyltransferase;PDBTitle: structure of atp-phosphoribosyltransferase from e. coli complexed with2 pr-atp
119 d1z7me1
not modelled
23.4
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like120 c3uifA_
not modelled
23.4
19
PDB header: transport proteinChain: A: PDB Molecule: sulfonate abc transporter, periplasmic sulfonate-bindingPDBTitle: crystal structure of putative sulfonate abc transporter, periplasmic2 sulfonate-binding protein ssua from methylobacillus flagellatus kt