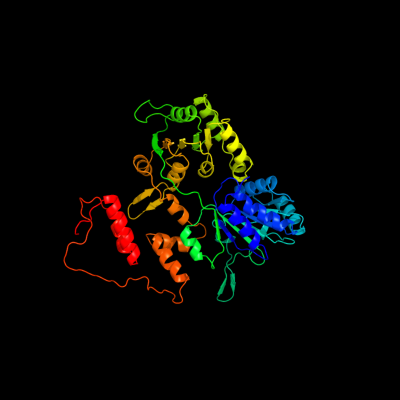
1 c2eyqA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: transcription-repair coupling factor;PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
2 c1gm5A_
100.0
19
PDB header: helicaseChain: A: PDB Molecule: recg;PDBTitle: structure of recg bound to three-way dna junction
3 c2xgjA_
100.0
17
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: structure of mtr4, a dexh helicase involved in nuclear rna2 processing and surveillance
4 c4a4zA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: antiviral helicase ski2;PDBTitle: crystal structure of the s. cerevisiae dexh helicase ski2 bound to2 amppnp
5 c2v1xB_
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent dna helicase q1;PDBTitle: crystal structure of human recq-like dna helicase
6 c1oywA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase;PDBTitle: structure of the recq catalytic core
7 c3l9oA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: crystal structure of mtr4, a co-factor of the nuclear exosome
8 c2va8A_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: ski2-type helicase;PDBTitle: dna repair helicase hel308
9 c1gl9B_
100.0
18
PDB header: topoisomeraseChain: B: PDB Molecule: reverse gyrase;PDBTitle: archaeoglobus fulgidus reverse gyrase complexed with adpnp
10 c3bxzA_
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: preprotein translocase subunit seca;PDBTitle: crystal structure of the isolated dead motor domains from2 escherichia coli seca
11 c2zj2A_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: putative ski2-type helicase;PDBTitle: archaeal dna helicase hjm apo state in form 1
12 c1c4oA_
100.0
20
PDB header: replicationChain: A: PDB Molecule: dna nucleotide excision repair enzyme uvrb;PDBTitle: crystal structure of the dna nucleotide excision repair enzyme uvrb2 from thermus thermophilus
13 c2p6uA_
100.0
18
PDB header: dna binding proteinChain: A: PDB Molecule: afuhel308 helicase;PDBTitle: apo structure of the hel308 superfamily 2 helicase
14 c3i5yA_
100.0
20
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase mss116;PDBTitle: structure of mss116p bound to ssrna containing a single 5-bru and amp-2 pnp
15 c2db3D_
100.0
19
PDB header: hydrolase/rnaChain: D: PDB Molecule: atp-dependent rna helicase vasa;PDBTitle: structural basis for rna unwinding by the dead-box protein2 drosophila vasa
16 c3tmiA_
100.0
15
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase ddx58;PDBTitle: structural basis for rna recognition and activation of rig-i
17 c1hv8B_
100.0
21
PDB header: rna binding proteinChain: B: PDB Molecule: putative atp-dependent rna helicase mj0669;PDBTitle: crystal structure of a dead box protein from the2 hyperthermophile methanococcus jannaschii
18 c3rc8A_
100.0
18
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase supv3l1, mitochondrial;PDBTitle: human mitochondrial helicase suv3 in complex with short rna fragment
19 c2d7dA_
100.0
16
PDB header: hydrolase/dnaChain: A: PDB Molecule: uvrabc system protein b;PDBTitle: structural insights into the cryptic dna dependent atp-ase2 activity of uvrb
20 c3ewsA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase ddx19b;PDBTitle: human dead-box rna-helicase ddx19 in complex with adp
21 c1wp9D_
not modelled
100.0
19
PDB header: hydrolaseChain: D: PDB Molecule: atp-dependent rna helicase, putative;PDBTitle: crystal structure of pyrococcus furiosus hef helicase domain
22 c3kx2A_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: pre-mrna-splicing factor atp-dependent rnaPDBTitle: crystal structure of prp43p in complex with adp
23 c2ocaA_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase uvsw;PDBTitle: the crystal structure of t4 uvsw
24 c2jlrA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: serine protease subunit ns3;PDBTitle: dengue virus 4 ns3 helicase in complex with amppnp
25 c1s2mA_
not modelled
100.0
16
PDB header: rna binding proteinChain: A: PDB Molecule: putative atp-dependent rna helicase dhh1;PDBTitle: crystal structure of the dead box protein dhh1p
26 c3fhtA_
not modelled
100.0
17
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase ddx19b;PDBTitle: crystal structure of human dbp5 in complex with amppnp and rna
27 c1xtkA_
not modelled
100.0
17
PDB header: gene regulationChain: A: PDB Molecule: probable atp-dependent rna helicase p47;PDBTitle: structure of decd to dead mutation of human uap56
28 c2vbcA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: dengue 4 ns3 full-length protein;PDBTitle: crystal structure of the ns3 protease-helicase from dengue2 virus
29 c2z0mA_
not modelled
100.0
23
PDB header: rna binding proteinChain: A: PDB Molecule: 337aa long hypothetical atp-dependent rnaPDBTitle: crystal structure of hypothetical atp-dependent rna2 helicase from sulfolobus tokodaii
30 c3pexA_
not modelled
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dbp5;PDBTitle: s. cerevisiae dbp5 l327v bound to gle1 h337r and ip6
31 c3oiyB_
not modelled
100.0
15
PDB header: isomeraseChain: B: PDB Molecule: reverse gyrase helicase domain;PDBTitle: helicase domain of reverse gyrase from thermotoga maritima
32 c2v6jA_
not modelled
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: rna helicase;PDBTitle: kokobera virus helicase: mutant met47thr
33 c2fwrA_
not modelled
100.0
14
PDB header: dna binding proteinChain: A: PDB Molecule: dna repair protein rad25;PDBTitle: structure of archaeoglobus fulgidis xpb
34 c2w74B_
not modelled
100.0
13
PDB header: hydrolaseChain: B: PDB Molecule: type i restriction enzyme ecor124ii r protein;PDBTitle: mutant (k220r) of the hsdr subunit of the ecor124i2 restriction enzyme in complex with atp
35 c2hxyC_
not modelled
100.0
17
PDB header: hydrolaseChain: C: PDB Molecule: probable atp-dependent rna helicase ddx48;PDBTitle: crystal structure of human apo-eif4aiii
36 c2wv9A_
not modelled
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: flavivirin protease ns2b regulatory subunit, flavivirinPDBTitle: crystal structure of the ns3 protease-helicase from murray2 valley encephalitis virus
37 c2z83A_
not modelled
100.0
20
PDB header: viral proteinChain: A: PDB Molecule: helicase/nucleoside triphosphatase;PDBTitle: crystal structure of catalytic domain of japanese2 encephalitis virus ns3 helicase/nucleoside triphosphatase3 at a resolution 1.8
38 c2fsgA_
not modelled
100.0
16
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase seca subunit;PDBTitle: complex seca:atp from escherichia coli
39 c1ymfA_
not modelled
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: genome polyprotein [contains: flavivirinPDBTitle: crystal structure of yellow fever virus ns3 helicase2 complexed with adp
40 c2vsxA_
not modelled
100.0
17
PDB header: translation/hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase eif4a;PDBTitle: crystal structure of a translation initiation complex
41 c2i4iA_
not modelled
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase ddx3x;PDBTitle: crystal structure of human dead-box rna helicase ddx3x
42 c3tbkA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: rig-i helicase domain;PDBTitle: mouse rig-i atpase domain
43 d2eyqa3
not modelled
100.0
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain44 c1a1vA_
not modelled
100.0
18
PDB header: hydrolase/dnaChain: A: PDB Molecule: protein (ns3 protein);PDBTitle: hepatitis c virus ns3 helicase domain complexed with single2 stranded sdna
45 c2qeqA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: flavivirin protease ns3 catalytic subunit;PDBTitle: crystal structure of kunjin virus ns3 helicase
46 d1gm5a3
not modelled
100.0
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain47 c3h1tA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: type i site-specific restriction-modificationPDBTitle: the fragment structure of a putative hsdr subunit of a type2 i restriction enzyme from vibrio vulnificus yj016
48 c1tf2A_
not modelled
100.0
18
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of seca:adp in an open conformation from2 bacillus subtilis
49 c2f55C_
not modelled
100.0
18
PDB header: hydrolase/dnaChain: C: PDB Molecule: polyprotein;PDBTitle: two hepatitis c virus ns3 helicase domains complexed with2 the same strand of dna
50 c3dl8B_
not modelled
100.0
20
PDB header: protein transportChain: B: PDB Molecule: protein translocase subunit seca;PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
51 c2d7gD_
100.0
100
PDB header: hydrolaseChain: D: PDB Molecule: primosomal protein n;PDBTitle: crystal structure of the aa complex of the n-terminal2 domain of pria
52 d2bmfa2
not modelled
100.0
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RNA helicase53 c3mwyW_
not modelled
99.9
15
PDB header: hydrolaseChain: W: PDB Molecule: chromo domain-containing protein 1;PDBTitle: crystal structure of the chromodomain-atpase portion of the yeast chd12 chromatin remodeler
54 c1z3iX_
not modelled
99.9
15
PDB header: recombination/dna bindingChain: X: PDB Molecule: similar to rad54-like;PDBTitle: structure of the swi2/snf2 chromatin remodeling domain of eukaryotic2 rad54
55 c1cu1B_
not modelled
99.9
18
PDB header: hydrolaseChain: B: PDB Molecule: protein (protease/helicase ns3);PDBTitle: crystal structure of an enzyme complex from hepatitis c2 virus
56 c2w00B_
not modelled
99.9
14
PDB header: hydrolaseChain: B: PDB Molecule: hsdr;PDBTitle: crystal structure of the hsdr subunit of the ecor124i2 restriction enzyme in complex with atp
57 c3dmqA_
not modelled
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: rna polymerase-associated protein rapa;PDBTitle: crystal structure of rapa, a swi2/snf2 protein that2 recycles rna polymerase during transcription
58 c3crw1_
not modelled
99.9
16
PDB header: hydrolaseChain: 1: PDB Molecule: xpd/rad3 related dna helicase;PDBTitle: "xpd_apo"
59 c1z63A_
not modelled
99.9
13
PDB header: hydrolase/dna complexChain: A: PDB Molecule: helicase of the snf2/rad54 hamily;PDBTitle: sulfolobus solfataricus swi2/snf2 atpase core in complex2 with dsdna
60 d1oywa2
not modelled
99.9
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain61 c3dkpA_
not modelled
99.9
18
PDB header: hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx52;PDBTitle: human dead-box rna-helicase ddx52, conserved domain i in complex with2 adp
62 c3fe2B_
not modelled
99.9
18
PDB header: hydrolaseChain: B: PDB Molecule: probable atp-dependent rna helicase ddx5;PDBTitle: human dead-box rna helicase ddx5 (p68), conserved domain i in complex2 with adp
63 c3ly5A_
not modelled
99.9
24
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase ddx18;PDBTitle: ddx18 dead-domain
64 d2eyqa5
not modelled
99.9
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain65 d1gl9b1
not modelled
99.9
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Helicase-like "domain" of reverse gyrase66 c2vl7A_
not modelled
99.9
16
PDB header: unknown functionChain: A: PDB Molecule: xpd;PDBTitle: structure of s. tokodaii xpd4
67 d1gkub1
not modelled
99.9
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Helicase-like "domain" of reverse gyrase68 c2pl3A_
not modelled
99.9
21
PDB header: hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx10;PDBTitle: human dead-box rna helicase ddx10, dead domain in complex with adp
69 c2kbeA_
not modelled
99.8
18
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dbp5;PDBTitle: solution structure of amino-terminal domain of dbp5p
70 d1s2ma1
not modelled
99.8
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain71 d1hv8a1
not modelled
99.8
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain72 d2j0sa1
not modelled
99.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain73 d1q0ua_
not modelled
99.8
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain74 d1rifa_
not modelled
99.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: DNA helicase UvsW75 c2gxqA_
not modelled
99.8
21
PDB header: hydrolaseChain: A: PDB Molecule: heat resistant rna dependent atpase;PDBTitle: hera n-terminal domain in complex with amp, crystal form 1
76 c3berA_
not modelled
99.8
21
PDB header: hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx47;PDBTitle: human dead-box rna-helicase ddx47, conserved domain i in complex with2 amp
77 c3iuyB_
not modelled
99.8
19
PDB header: hydrolaseChain: B: PDB Molecule: probable atp-dependent rna helicase ddx53;PDBTitle: crystal structure of ddx53 dead-box domain
78 d2p6ra3
not modelled
99.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain79 d2fwra2
not modelled
99.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain80 c3b6eA_
not modelled
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: interferon-induced helicase c domain-containing protein 1;PDBTitle: crystal structure of human dech-box rna helicase mda5 (melanoma2 differentiation-associated protein 5), dech-domain
81 d1wp9a1
not modelled
99.8
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain82 c2oxcA_
not modelled
99.8
15
PDB header: hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx20;PDBTitle: human dead-box rna helicase ddx20, dead domain in complex2 with adp
83 d1gm5a4
not modelled
99.8
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain84 d1wrba1
not modelled
99.8
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain85 d1qdea_
not modelled
99.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain86 d1a1va1
not modelled
99.8
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RNA helicase87 d2fz4a1
not modelled
99.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain88 d1veca_
not modelled
99.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain89 c3llmB_
not modelled
99.8
19
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase a;PDBTitle: crystal structure analysis of a rna helicase
90 d1t5la2
not modelled
99.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain91 d1t6na_
not modelled
99.8
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain92 d1oywa3
not modelled
99.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain93 c3fmoB_
not modelled
99.8
22
PDB header: oncoprotein/hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase ddx19b;PDBTitle: crystal structure of the nucleoporin nup214 in complex with the dead-2 box helicase ddx19
94 d1c4oa2
not modelled
99.7
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain95 c3fhcB_
not modelled
99.7
21
PDB header: transport protein/hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase ddx19b;PDBTitle: crystal structure of human dbp5 in complex with nup214
96 d1nkta3
not modelled
99.7
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain97 d1tf5a3
not modelled
99.7
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain98 d2g9na1
not modelled
99.7
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain99 c3fmpD_
not modelled
99.7
22
PDB header: oncoprotein/hydrolaseChain: D: PDB Molecule: atp-dependent rna helicase ddx19b;PDBTitle: crystal structure of the nucleoporin nup214 in complex with the dead-2 box helicase ddx19
100 d1yksa1
not modelled
99.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RNA helicase101 d2j0sa2
not modelled
99.7
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain102 c2vsfA_
not modelled
99.7
19
PDB header: hydrolaseChain: A: PDB Molecule: dna repair helicase rad3 related protein;PDBTitle: structure of xpd from thermoplasma acidophilum
103 c2vdaA_
not modelled
99.7
20
PDB header: protein transportChain: A: PDB Molecule: translocase subunit seca;PDBTitle: solution structure of the seca-signal peptide complex
104 d1hv8a2
not modelled
99.7
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain105 d1c4oa1
not modelled
99.7
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain106 c1nl3B_
not modelled
99.6
20
PDB header: protein transportChain: B: PDB Molecule: preprotein translocase seca 1 subunit;PDBTitle: crystal structure of the seca protein translocation atpase2 from mycobacterium tuberculosis in apo form
107 d1s2ma2
not modelled
99.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain108 c3i32A_
not modelled
99.6
26
PDB header: rna binding protein,hydrolaseChain: A: PDB Molecule: heat resistant rna dependent atpase;PDBTitle: dimeric structure of a hera helicase fragment including the c-terminal2 reca domain, the dimerization domain, and the rna binding domain
109 c3juxA_
not modelled
99.6
22
PDB header: protein transportChain: A: PDB Molecule: protein translocase subunit seca;PDBTitle: structure of the translocation atpase seca from thermotoga2 maritima
110 d2p6ra4
not modelled
99.6
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain111 c2hjvB_
not modelled
99.6
22
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase dbpa;PDBTitle: structure of the second domain (residues 207-368) of the2 bacillus subtilis yxin protein
112 c2p6nA_
not modelled
99.6
15
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase ddx41;PDBTitle: human dead-box rna helicase ddx41, helicase domain
113 c2ipcB_
not modelled
99.6
19
PDB header: transport proteinChain: B: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of the translocation atpase seca from thermus2 thermophilus reveals a parallel, head-to-head dimer
114 c3dinB_
not modelled
99.6
22
PDB header: membrane protein, protein transportChain: B: PDB Molecule: protein translocase subunit seca;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
115 c3earA_
not modelled
99.5
27
PDB header: hydrolaseChain: A: PDB Molecule: hera;PDBTitle: novel dimerization motif in the dead box rna helicase hera: form 1,2 partial dimer
116 d2g2ja1
not modelled
99.5
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain117 d1t5ia_
not modelled
99.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain118 d1wp9a2
not modelled
99.5
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain119 d1z3ix2
not modelled
99.4
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain120 d1nkta4
not modelled
99.4
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain