| 1 | d1a99a_ |
|
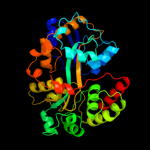 |
100.0 |
100 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 2 | d1pota_ |
|
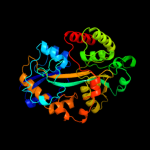 |
100.0 |
36 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 3 | c2v84A_ |
|
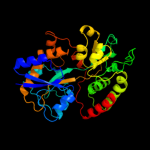 |
100.0 |
25 |
PDB header:transport protein
Chain: A: PDB Molecule:spermidine/putrescine abc transporter, periplasmic
PDBTitle: crystal structure of the tp0655 (tppotd) lipoprotein of2 treponema pallidum
|
| 4 | c3osqA_ |
|
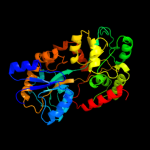 |
100.0 |
20 |
PDB header:fluorescent protein, transport protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein, green fluorescent
PDBTitle: maltose-bound maltose sensor engineered by insertion of circularly2 permuted green fluorescent protein into e. coli maltose binding3 protein at position 175
|
| 5 | c3h4zC_ |
|
 |
100.0 |
18 |
PDB header:allergen
Chain: C: PDB Molecule:maltose-binding periplasmic protein fused with allergen
PDBTitle: crystal structure of an mbp-der p 7 fusion protein
|
| 6 | c3f5fA_ |
|
 |
100.0 |
18 |
PDB header:transport, transferase
Chain: A: PDB Molecule:maltose-binding periplasmic protein, heparan
PDBTitle: crystal structure of heparan sulfate 2-o-sulfotransferase2 from gallus gallus as a maltose binding protein fusion.
|
| 7 | c3o3uN_ |
|
 |
100.0 |
17 |
PDB header:transport protein, signaling protein
Chain: N: PDB Molecule:maltose-binding periplasmic protein, advanced glycosylation
PDBTitle: crystal structure of human receptor for advanced glycation endproducts2 (rage)
|
| 8 | c3ob4A_ |
|
 |
100.0 |
18 |
PDB header:allergen
Chain: A: PDB Molecule:maltose abc transporter periplasmic protein, arah 2;
PDBTitle: mbp-fusion protein of the major peanut allergen ara h 2
|
| 9 | c3py7A_ |
|
 |
100.0 |
18 |
PDB header:viral protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein,paxillin ld1,protein e6
PDBTitle: crystal structure of full-length bovine papillomavirus oncoprotein e62 in complex with ld1 motif of paxillin at 2.3a resolution
|
| 10 | c3d4cA_ |
|
 |
100.0 |
17 |
PDB header:cell adhesion
Chain: A: PDB Molecule:maltose-binding periplasmic protein, linker, zona pellucida
PDBTitle: zp-n domain of mammalian sperm receptor zp3 (crystal form i)
|
| 11 | c3dm0A_ |
|
 |
100.0 |
18 |
PDB header:sugar binding protein,signaling protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein fused with
PDBTitle: maltose binding protein fusion with rack1 from a. thaliana
|
| 12 | c3c4mA_ |
|
 |
100.0 |
17 |
PDB header:membrane protein
Chain: A: PDB Molecule:fusion protein of maltose-binding periplasmic protein and
PDBTitle: structure of human parathyroid hormone in complex with the2 extracellular domain of its g-protein-coupled receptor (pth1r)
|
| 13 | c3mp6A_ |
|
 |
100.0 |
18 |
PDB header:histone binding protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein, linker, saga-
PDBTitle: complex structure of sgf29 and dimethylated h3k4
|
| 14 | c2vgqA_ |
|
 |
100.0 |
18 |
PDB header:immune system/transport
Chain: A: PDB Molecule:maltose-binding periplasmic protein,
PDBTitle: crystal structure of human ips-1 card
|
| 15 | c1y4cA_ |
|
 |
100.0 |
18 |
PDB header:de novo protein
Chain: A: PDB Molecule:maltose binding protein fused with designed
PDBTitle: designed helical protein fusion mbp
|
| 16 | c1r6zA_ |
|
 |
100.0 |
18 |
PDB header:gene regulation
Chain: A: PDB Molecule:chimera of maltose-binding periplasmic protein and
PDBTitle: the crystal structure of the argonaute2 paz domain (as a mbp fusion)
|
| 17 | c3oaiB_ |
|
 |
100.0 |
17 |
PDB header:membrane protein, cell adhesion
Chain: B: PDB Molecule:maltose-binding periplasmic protein, myelin protein p0;
PDBTitle: crystal structure of the extra-cellular domain of human myelin protein2 zero
|
| 18 | c1hsjA_ |
|
 |
100.0 |
17 |
PDB header:transcription/sugar binding protein
Chain: A: PDB Molecule:fusion protein consisting of staphylococcus
PDBTitle: sarr mbp fusion structure
|
| 19 | c3csgA_ |
|
 |
100.0 |
18 |
PDB header:de novo protein, sugar binding protein
Chain: A: PDB Molecule:maltose-binding protein monobody ys1 fusion;
PDBTitle: crystal structure of monobody ys1(mbp-74)/maltose binding2 protein fusion complex
|
| 20 | d1eu8a_ |
|
 |
100.0 |
15 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 21 | c3k02A_ |
|
not modelled |
100.0 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:acarbose/maltose binding protein gach;
PDBTitle: crystal structures of the gach receptor of streptomyces glaucescens2 gla.o in the unliganded form and in complex with acarbose and an3 acarbose homolog. comparison with acarbose-loaded maltose binding4 protein of salmonella typhimurium.
|
| 22 | c2nvuB_ |
|
not modelled |
100.0 |
18 |
PDB header:protein turnover, ligase
Chain: B: PDB Molecule:maltose binding protein/nedd8-activating enzyme
PDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
|
| 23 | c3osrA_ |
|
not modelled |
100.0 |
18 |
PDB header:fluorescent protein, transport protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein, green fluorescent
PDBTitle: maltose-bound maltose sensor engineered by insertion of circularly2 permuted green fluorescent protein into e. coli maltose binding3 protein at position 311
|
| 24 | c3uorB_ |
|
not modelled |
100.0 |
13 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:abc transporter sugar binding protein;
PDBTitle: the structure of the sugar-binding protein male from the phytopathogen2 xanthomonas citri
|
| 25 | c3qufB_ |
|
not modelled |
100.0 |
13 |
PDB header:transport protein
Chain: B: PDB Molecule:extracellular solute-binding protein, family 1;
PDBTitle: the structure of a family 1 extracellular solute-binding protein from2 bifidobacterium longum subsp. infantis
|
| 26 | d1elja_ |
|
not modelled |
100.0 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 27 | c3ehuA_ |
|
not modelled |
100.0 |
18 |
PDB header:membrane protein
Chain: A: PDB Molecule:fusion protein of crfr1 extracellular domain and mbp;
PDBTitle: crystal structure of the extracellular domain of human corticotropin2 releasing factor receptor type 1 (crfr1) in complex with crf
|
| 28 | c2zykA_ |
|
not modelled |
100.0 |
14 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:solute-binding protein;
PDBTitle: crystal structure of cyclo/maltodextrin-binding protein2 complexed with gamma-cyclodextrin
|
| 29 | c1mg1A_ |
|
not modelled |
100.0 |
17 |
PDB header:viral protein
Chain: A: PDB Molecule:protein (htlv-1 gp21 ectodomain/maltose-binding protein
PDBTitle: htlv-1 gp21 ectodomain/maltose-binding protein chimera
|
| 30 | c2xd3A_ |
|
not modelled |
100.0 |
15 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:maltose/maltodextrin-binding protein;
PDBTitle: the crystal structure of malx from streptococcus pneumoniae2 in complex with maltopentaose.
|
| 31 | c3iouB_ |
|
not modelled |
100.0 |
18 |
PDB header:signaling protein
Chain: B: PDB Molecule:maltose-binding protein, huntingtin fusion
PDBTitle: huntingtin amino-terminal region with 17 gln residues -2 crystal c94
|
| 32 | c3oo6A_ |
|
not modelled |
100.0 |
11 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:abc transporter binding protein acbh;
PDBTitle: crystal structures and biochemical characterization of the bacterial2 solute receptor acbh reveal an unprecedented exclusive substrate3 preference for b-d-galactopyranose
|
| 33 | c1svxB_ |
|
not modelled |
100.0 |
18 |
PDB header:de novo protein/sugar binding protein
Chain: B: PDB Molecule:maltose-binding periplasmic protein;
PDBTitle: crystal structure of a designed selected ankyrin repeat2 protein in complex with the maltose binding protein
|
| 34 | c1mh3A_ |
|
not modelled |
100.0 |
17 |
PDB header:sugar binding, dna binding protein
Chain: A: PDB Molecule:maltose binding-a1 homeodomain protein chimera;
PDBTitle: maltose binding-a1 homeodomain protein chimera, crystal2 form i
|
| 35 | c2gh9A_ |
|
not modelled |
100.0 |
13 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:maltose/maltodextrin-binding protein;
PDBTitle: thermus thermophilus maltotriose binding protein bound with2 maltotriose
|
| 36 | d1laxa_ |
|
not modelled |
100.0 |
16 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 37 | c3rpwA_ |
|
not modelled |
100.0 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter;
PDBTitle: the crystal structure of an abc transporter from rhodopseudomonas2 palustris cga009
|
| 38 | c2b3fD_ |
|
not modelled |
100.0 |
14 |
PDB header:sugar binding protein
Chain: D: PDB Molecule:glucose-binding protein;
PDBTitle: thermus thermophilus glucose/galactose binding protein2 bound with galactose
|
| 39 | c2fncA_ |
|
not modelled |
100.0 |
15 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:maltose abc transporter, periplasmic maltose-binding
PDBTitle: thermotoga maritima maltotriose binding protein bound with2 maltotriose.
|
| 40 | c2uvgA_ |
|
not modelled |
100.0 |
15 |
PDB header:sugar-binding protein
Chain: A: PDB Molecule:abc type periplasmic sugar-binding protein;
PDBTitle: structure of a periplasmic oligogalacturonide binding2 protein from yersinia enterocolitica
|
| 41 | c2z8fB_ |
|
not modelled |
100.0 |
13 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:galacto-n-biose/lacto-n-biose i transporter substrate-
PDBTitle: the galacto-n-biose-/lacto-n-biose i-binding protein (gl-bp) of the2 abc transporter from bifidobacterium longum in complex with lacto-n-3 tetraose
|
| 42 | d1ursa_ |
|
not modelled |
100.0 |
11 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 43 | c1ursA_ |
|
not modelled |
100.0 |
11 |
PDB header:maltose-binding protein
Chain: A: PDB Molecule:maltose-binding protein;
PDBTitle: x-ray structures of the maltose-maltodextrin binding2 protein of the thermoacidophilic bacterium alicyclobacillus3 acidocaldarius
|
| 44 | c2i58B_ |
|
not modelled |
100.0 |
20 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:sugar abc transporter, sugar-binding protein;
PDBTitle: crystal structure of rafe from streptococcus pneumoniae complexed with2 raffinose
|
| 45 | c3i3vC_ |
|
not modelled |
100.0 |
12 |
PDB header:transport protein
Chain: C: PDB Molecule:probable secreted solute-binding lipoprotein;
PDBTitle: crystal structure of probable secreted solute-binding2 lipoprotein from streptomyces coelicolor
|
| 46 | d3thia_ |
|
not modelled |
100.0 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 47 | c2w7yA_ |
|
not modelled |
100.0 |
12 |
PDB header:sugar-binding protein
Chain: A: PDB Molecule:probable sugar abc transporter, sugar-binding
PDBTitle: structure of a streptococcus pneumoniae solute-binding2 protein in complex with the blood group a-trisaccharide.
|
| 48 | d1y4ta_ |
|
not modelled |
100.0 |
16 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 49 | c3pu5A_ |
|
not modelled |
100.0 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular solute-binding protein;
PDBTitle: the crystal structure of a putative extracellular solute-binding2 protein from bordetella parapertussis
|
| 50 | c2qryD_ |
|
not modelled |
100.0 |
15 |
PDB header:transport protein
Chain: D: PDB Molecule:thiamine-binding periplasmic protein;
PDBTitle: periplasmic thiamin binding protein
|
| 51 | c3c9hB_ |
|
not modelled |
100.0 |
13 |
PDB header:transport protein
Chain: B: PDB Molecule:abc transporter, substrate binding protein;
PDBTitle: crystal structure of the substrate binding protein of the abc2 transporter from agrobacterium tumefaciens
|
| 52 | d1q35a_ |
|
not modelled |
100.0 |
17 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 53 | c2pt1A_ |
|
not modelled |
100.0 |
16 |
PDB header:metal transport
Chain: A: PDB Molecule:iron transport protein;
PDBTitle: futa1 synechocystis pcc 6803
|
| 54 | d1y9ua_ |
|
not modelled |
100.0 |
15 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 55 | d1xvxa_ |
|
not modelled |
100.0 |
18 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 56 | c2vozA_ |
|
not modelled |
100.0 |
18 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:periplasmic iron-binding protein;
PDBTitle: apo futa2 from synechocystis pcc6803
|
| 57 | d1xc1a_ |
|
not modelled |
100.0 |
17 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 58 | d1nnfa_ |
|
not modelled |
100.0 |
17 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 59 | d1xvya_ |
|
not modelled |
100.0 |
18 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 60 | d1j1na_ |
|
not modelled |
100.0 |
15 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 61 | d1y3na1 |
|
not modelled |
100.0 |
17 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 62 | c3ombA_ |
|
not modelled |
100.0 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular solute-binding protein, family 1;
PDBTitle: crystal structure of extracellular solute-binding protein from2 bifidobacterium longum subsp. infantis
|
| 63 | d2onsa1 |
|
not modelled |
100.0 |
8 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 64 | c3cfxA_ |
|
not modelled |
100.0 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein ma_0280;
PDBTitle: crystal structure of m. acetivorans periplasmic binding protein2 moda/wtpa with bound tungstate
|
| 65 | c3k6wA_ |
|
not modelled |
99.9 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:solute-binding protein ma_0280;
PDBTitle: apo and ligand bound structures of moda from the archaeon2 methanosarcina acetivorans
|
| 66 | c3cfzA_ |
|
not modelled |
99.9 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein mj1186;
PDBTitle: crystal structure of m. jannaschii periplasmic binding2 protein moda/wtpa with bound tungstate
|
| 67 | c3cg3A_ |
|
not modelled |
99.9 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein ph0151;
PDBTitle: crystal structure of p. horikoshii periplasmic binding2 protein moda/wtpa with bound tungstate
|
| 68 | c3cg1A_ |
|
not modelled |
99.9 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein pf0080;
PDBTitle: crystal structure of p. furiosus periplasmic binding protein2 moda/wtpa with bound tungstate
|
| 69 | d1atga_ |
|
not modelled |
99.9 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 70 | d1sbpa_ |
|
not modelled |
99.9 |
16 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 71 | c2h5yC_ |
|
not modelled |
99.9 |
12 |
PDB header:metal transport
Chain: C: PDB Molecule:molybdate-binding periplasmic protein;
PDBTitle: crystallographic structure of the molybdate-binding protein of2 xanthomonas citri at 1.7 ang resolution bound to molybdate
|
| 72 | d1amfa_ |
|
not modelled |
99.8 |
16 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 73 | c3fj7A_ |
|
not modelled |
99.8 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:major antigenic peptide peb3;
PDBTitle: crystal structure of l-phospholactate bound peb3
|
| 74 | c3lr1A_ |
|
not modelled |
99.7 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:tungstate abc transporter, periplasmic tungstate-
PDBTitle: the crystal structure of the tungstate abc transporter from2 geobacter sulfurreducens
|
| 75 | c3muqB_ |
|
not modelled |
99.6 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the crystal structure of a conserved functionally unknown protein from2 vibrio parahaemolyticus rimd 2210633
|
| 76 | c3kn3C_ |
|
not modelled |
99.5 |
13 |
PDB header:transcription
Chain: C: PDB Molecule:putative periplasmic protein;
PDBTitle: crystal structure of lysr substrate binding domain (25-263) of2 putative periplasmic protein from wolinella succinogenes
|
| 77 | c1twyG_ |
|
not modelled |
99.3 |
13 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:abc transporter, periplasmic substrate-binding protein;
PDBTitle: crystal structure of an abc-type phosphate transport receptor from2 vibrio cholerae
|
| 78 | d1twya_ |
|
not modelled |
99.3 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 79 | d1pc3a_ |
|
not modelled |
97.4 |
15 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 80 | d1xs5a_ |
|
not modelled |
96.6 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 81 | c3tqwA_ |
|
not modelled |
96.1 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:methionine-binding protein;
PDBTitle: structure of a abc transporter, periplasmic substrate-binding protein2 from coxiella burnetii
|
| 82 | c3k2dA_ |
|
not modelled |
95.7 |
10 |
PDB header:immune system
Chain: A: PDB Molecule:abc-type metal ion transport system, periplasmic component;
PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
|
| 83 | c3e4rA_ |
|
not modelled |
94.2 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein;
PDBTitle: crystal structure of the alkanesulfonate binding protein2 (ssua) from the phytopathogenic bacteria xanthomonas3 axonopodis pv. citri bound to hepes
|
| 84 | c3gxaA_ |
|
not modelled |
93.2 |
12 |
PDB header:protein binding
Chain: A: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of gna1946
|
| 85 | c3ir1F_ |
|
not modelled |
93.2 |
13 |
PDB header:protein binding
Chain: F: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
|
| 86 | c3cvgC_ |
|
not modelled |
93.0 |
19 |
PDB header:metal binding protein
Chain: C: PDB Molecule:putative metal binding protein;
PDBTitle: crystal structure of a periplasmic putative metal binding protein
|
| 87 | c3n5lA_ |
|
not modelled |
91.5 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:binding protein component of abc phosphonate transporter;
PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
|
| 88 | c3tmgA_ |
|
not modelled |
89.8 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine, l-proline abc transporter,
PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
|
| 89 | d1ixha_ |
|
not modelled |
81.6 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 90 | c2x26A_ |
|
not modelled |
80.3 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic aliphatic sulphonates-binding protein;
PDBTitle: crystal structure of the periplasmic aliphatic sulphonate2 binding protein ssua from escherichia coli
|
| 91 | c2q2aD_ |
|
not modelled |
78.5 |
12 |
PDB header:transport protein
Chain: D: PDB Molecule:artj;
PDBTitle: crystal structures of the arginine-, lysine-, histidine-2 binding protein artj from the thermophilic bacterium3 geobacillus stearothermophilus
|
| 92 | c3l6gA_ |
|
not modelled |
74.8 |
13 |
PDB header:glycine betaine-binding protein
Chain: A: PDB Molecule:betaine abc transporter permease and substrate binding
PDBTitle: crystal structure of lactococcal opuac in its open conformation
|
| 93 | d1p99a_ |
|
not modelled |
71.8 |
11 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 94 | c1p99A_ |
|
not modelled |
71.8 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pg110;
PDBTitle: 1.7a crystal structure of protein pg110 from staphylococcus2 aureus
|
| 95 | d2v3qa1 |
|
not modelled |
50.6 |
8 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 96 | c2rejA_ |
|
not modelled |
49.5 |
14 |
PDB header:choline-binding protein
Chain: A: PDB Molecule:putative glycine betaine abc transporter protein;
PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
|
| 97 | c3ix1A_ |
|
not modelled |
39.0 |
12 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding
PDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
|
| 98 | c3ix1B_ |
|
not modelled |
39.0 |
12 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding
PDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
|
| 99 | c2f78A_ |
|
not modelled |
34.4 |
11 |
PDB header:gene regulation
Chain: A: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: benm effector binding domain with its effector benzoate
|
| 100 | c2h9qC_ |
|
not modelled |
33.3 |
15 |
PDB header:transcription
Chain: C: PDB Molecule:hth-type transcriptional regulator catm;
PDBTitle: crystal structure of the effector binding domain of a catm2 variant (r156h)
|
| 101 | c1jrjA_ |
|
not modelled |
32.0 |
17 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:exendin-4;
PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
|
| 102 | c2h9bB_ |
|
not modelled |
31.2 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: crystal structure of the effector binding domain of a benm variant2 (benm r156h/t157s)
|
| 103 | c1d0rA_ |
|
not modelled |
28.2 |
25 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:glucagon-like peptide-1-(7-36)-amide;
PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
|
| 104 | c3jv9B_ |
|
not modelled |
27.7 |
4 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, lysr family;
PDBTitle: the structure of a reduced form of oxyr from n. meningitidis
|
| 105 | c3un6A_ |
|
not modelled |
25.1 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein saouhsc_00137;
PDBTitle: 2.0 angstrom crystal structure of ligand binding component of abc-type2 import system from staphylococcus aureus with zinc bound
|
| 106 | c2ql3G_ |
|
not modelled |
24.1 |
13 |
PDB header:transcription
Chain: G: PDB Molecule:probable transcriptional regulator, lysr family protein;
PDBTitle: crystal structure of the c-terminal domain of a probable lysr family2 transcriptional regulator from rhodococcus sp. rha1
|
| 107 | d1whqa_ |
|
not modelled |
22.3 |
24 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 108 | d1ixca2 |
|
not modelled |
22.0 |
22 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 109 | c2f7cA_ |
|
not modelled |
22.0 |
17 |
PDB header:gene regulation
Chain: A: PDB Molecule:hth-type transcriptional regulator catm;
PDBTitle: catm effector binding domain with its effector cis,cis-muconate
|
| 110 | c3mz1D_ |
|
not modelled |
21.6 |
17 |
PDB header:transcription regulator
Chain: D: PDB Molecule:putative transcriptional regulator;
PDBTitle: the crystal structure of a possible transcription regulator protein2 from sinorhizobium meliloti 1021
|
| 111 | c3n6uA_ |
|
not modelled |
21.3 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:lysr type regulator of tsambcd;
PDBTitle: effector binding domain of tsar in complex with its inducer p-2 toluenesulfonate
|

