1 c3iytG_
99.9
13
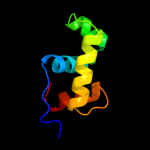
PDB header: apoptosisChain: G: PDB Molecule: apoptotic protease-activating factor 1;PDBTitle: structure of an apoptosome-procaspase-9 card complex
2 c1z6tC_
99.9
14
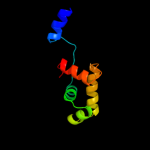
PDB header: apoptosisChain: C: PDB Molecule: apoptotic protease activating factor 1;PDBTitle: structure of the apoptotic protease-activating factor 12 bound to adp
3 c2q0oA_
99.9
19
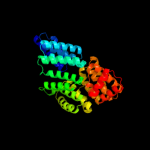
PDB header: transcriptionChain: A: PDB Molecule: probable transcriptional activator protein trar;PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
4 c3sztB_
99.9
25
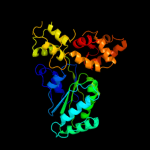
PDB header: transcriptionChain: B: PDB Molecule: quorum-sensing control repressor;PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
5 c1h0mD_
99.9
17
PDB header: transcription/dnaChain: D: PDB Molecule: transcriptional activator protein trar;PDBTitle: three-dimensional structure of the quorum sensing protein2 trar bound to its autoinducer and to its target dna
6 d1hz4a_
99.9
100
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Transcription factor MalT domain III7 c3qp5C_
99.9
29
PDB header: transcriptionChain: C: PDB Molecule: cvir transcriptional regulator;PDBTitle: crystal structure of cvir bound to antagonist chlorolactone (cl)
8 c2a5yB_
99.8
13
PDB header: apoptosisChain: B: PDB Molecule: ced-4;PDBTitle: structure of a ced-4/ced-9 complex
9 c2fnaA_
99.8
11
PDB header: atp-binding proteinChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of an archaeal aaa+ atpase (sso1545) from sulfolobus2 solfataricus p2 at 2.00 a resolution
10 c2qenA_
99.8
13
PDB header: unknown functionChain: A: PDB Molecule: walker-type atpase;PDBTitle: the walker-type atpase paby2304 of pyrococcus abyssi
11 d2fnaa2
99.8
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain12 c1zljE_
99.8
35
PDB header: transcriptionChain: E: PDB Molecule: dormancy survival regulator;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
13 c3c3wB_
99.8
36
PDB header: transcriptionChain: B: PDB Molecule: two component transcriptional regulatory protein devr;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
14 c3cloC_
99.8
30
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator containing a2 luxr dna binding domain (np_811094.1) from bacteroides3 thetaiotaomicron vpi-5482 at 2.04 a resolution
15 c2krfB_
99.8
26
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein coma;PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
16 c3klnC_
99.8
26
PDB header: transcriptionChain: C: PDB Molecule: transcriptional regulator, luxr family;PDBTitle: vibrio cholerae vpst
17 d1p4wa_
99.8
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)18 d1l3la1
99.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)19 d1fsea_
99.7
34
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)20 c1rnlA_
99.7
32
PDB header: signal transduction proteinChain: A: PDB Molecule: nitrate/nitrite response regulator protein narl;PDBTitle: the nitrate/nitrite response regulator protein narl from narl
21 c2rnjA_
not modelled
99.7
40
PDB header: transcriptionChain: A: PDB Molecule: response regulator protein vrar;PDBTitle: nmr structure of the s. aureus vrar dna binding domain
22 d1a04a1
not modelled
99.7
30
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)23 c1zn2A_
not modelled
99.7
45
PDB header: transcription regulatorChain: A: PDB Molecule: response regulatory protein;PDBTitle: low resolution structure of response regulator styr
24 c1x3uA_
not modelled
99.7
28
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein fixj;PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
25 c2jpcA_
not modelled
99.7
34
PDB header: dna binding proteinChain: A: PDB Molecule: ssrb;PDBTitle: ssrb dna binding protein
26 c1fnnB_
not modelled
99.7
17
PDB header: cell cycleChain: B: PDB Molecule: cell division control protein 6;PDBTitle: crystal structure of cdc6p from pyrobaculum aerophilum
27 c2qbyB_
not modelled
99.7
10
PDB header: replication/dnaChain: B: PDB Molecule: cell division control protein 6 homolog 3;PDBTitle: crystal structure of a heterodimer of cdc6/orc1 initiators2 bound to origin dna (from s. solfataricus)
28 d1yioa1
not modelled
99.7
45
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)29 c2v1uA_
not modelled
99.7
18
PDB header: replicationChain: A: PDB Molecule: cell division control protein 6 homolog;PDBTitle: structure of the aeropyrum pernix orc1 protein in complex2 with dna
30 c2qbyA_
not modelled
99.6
12
PDB header: replication/dnaChain: A: PDB Molecule: cell division control protein 6 homolog 1;PDBTitle: crystal structure of a heterodimer of cdc6/orc1 initiators2 bound to origin dna (from s. solfataricus)
31 d1fnna2
not modelled
99.4
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain32 c1w5sB_
not modelled
99.4
15
PDB header: replicationChain: B: PDB Molecule: origin recognition complex subunit 2 orc2;PDBTitle: structure of the aeropyrum pernix orc2 protein (adp form)
33 c1in8A_
not modelled
99.3
22
PDB header: dna binding proteinChain: A: PDB Molecule: holliday junction dna helicase ruvb;PDBTitle: thermotoga maritima ruvb t158v
34 c2chgB_
not modelled
99.2
12
PDB header: dna-binding proteinChain: B: PDB Molecule: replication factor c small subunit;PDBTitle: replication factor c domains 1 and 2
35 c3pvsA_
not modelled
99.1
16
PDB header: recombinationChain: A: PDB Molecule: replication-associated recombination protein a;PDBTitle: structure and biochemical activities of escherichia coli mgsa
36 c1sxjC_
not modelled
99.1
18
PDB header: replicationChain: C: PDB Molecule: activator 1 40 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader2 (replication factor c, rfc) bound to the dna sliding clamp3 (proliferating cell nuclear antigen, pcna)
37 c3pfiB_
not modelled
99.1
18
PDB header: hydrolaseChain: B: PDB Molecule: holliday junction atp-dependent dna helicase ruvb;PDBTitle: 2.7 angstrom resolution crystal structure of a probable holliday2 junction dna helicase (ruvb) from campylobacter jejuni subsp. jejuni3 nctc 11168 in complex with adenosine-5'-diphosphate
38 c2chvE_
not modelled
99.1
13
PDB header: dna-binding proteinChain: E: PDB Molecule: replication factor c small subunit;PDBTitle: replication factor c adpnp complex
39 d1w5sa2
not modelled
99.1
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain40 d1sxjd2
not modelled
99.0
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain41 d1in4a2
not modelled
99.0
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain42 c3te6A_
not modelled
99.0
12
PDB header: gene regulationChain: A: PDB Molecule: regulatory protein sir3;PDBTitle: crystal structure of the s. cerevisiae sir3 aaa+ domain
43 d1iqpa2
not modelled
99.0
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain44 c1iqpF_
not modelled
99.0
14
PDB header: replicationChain: F: PDB Molecule: rfcs;PDBTitle: crystal structure of the clamp loader small subunit from2 pyrococcus furiosus
45 d1sxjc2
not modelled
99.0
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain46 d1sxjb2
not modelled
99.0
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain47 d1r6bx2
not modelled
99.0
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain48 c1hqcB_
not modelled
98.9
22
PDB header: hydrolaseChain: B: PDB Molecule: ruvb;PDBTitle: structure of ruvb from thermus thermophilus hb8
49 c1sxjB_
not modelled
98.9
18
PDB header: replicationChain: B: PDB Molecule: activator 1 37 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader2 (replication factor c, rfc) bound to the dna sliding clamp3 (proliferating cell nuclear antigen, pcna)
50 c3u5zM_
not modelled
98.9
11
PDB header: dna binding protein/dnaChain: M: PDB Molecule: dna polymerase accessory protein 44;PDBTitle: structure of t4 bacteriophage clamp loader bound to the t4 clamp,2 primer-template dna, and atp analog
51 d1njfa_
not modelled
98.9
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain52 c1xwiA_
not modelled
98.9
16
PDB header: protein transportChain: A: PDB Molecule: skd1 protein;PDBTitle: crystal structure of vps4b
53 c2ce7B_
not modelled
98.9
17
PDB header: cell division proteinChain: B: PDB Molecule: cell division protein ftsh;PDBTitle: edta treated
54 d1sxja2
not modelled
98.9
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain55 c1sxjA_
not modelled
98.9
13
PDB header: replicationChain: A: PDB Molecule: activator 1 95 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader2 (replication factor c, rfc) bound to the dna sliding clamp3 (proliferating cell nuclear antigen, pcna)
56 d1a5ta2
not modelled
98.9
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain57 c3d8bB_
not modelled
98.9
17
PDB header: hydrolaseChain: B: PDB Molecule: fidgetin-like protein 1;PDBTitle: crystal structure of human fidgetin-like protein 1 in complex with adp
58 c1sxjD_
not modelled
98.9
12
PDB header: replicationChain: D: PDB Molecule: activator 1 41 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader2 (replication factor c, rfc) bound to the dna sliding clamp3 (proliferating cell nuclear antigen, pcna)
59 c1xxhB_
not modelled
98.9
14
PDB header: transferaseChain: B: PDB Molecule: dna polymerase iii subunit gamma;PDBTitle: atpgs bound e. coli clamp loader complex
60 c1sxjE_
not modelled
98.9
13
PDB header: replicationChain: E: PDB Molecule: activator 1 40 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader2 (replication factor c, rfc) bound to the dna sliding clamp3 (proliferating cell nuclear antigen, pcna)
61 d1sxje2
not modelled
98.9
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain62 d1qvra2
not modelled
98.9
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain63 d1ixsb2
not modelled
98.9
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain64 c3b9pA_
not modelled
98.8
18
PDB header: hydrolaseChain: A: PDB Molecule: cg5977-pa, isoform a;PDBTitle: spastin
65 c3bosA_
not modelled
98.8
13
PDB header: hydrolase regulator,dna binding proteinChain: A: PDB Molecule: putative dna replication factor;PDBTitle: crystal structure of a putative dna replication regulator hda2 (sama_1916) from shewanella amazonensis sb2b at 1.75 a resolution
66 c2zamA_
not modelled
98.8
18
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associating protein 4b;PDBTitle: crystal structure of mouse skd1/vps4b apo-form
67 c1r6bX_
not modelled
98.8
12
PDB header: hydrolaseChain: X: PDB Molecule: clpa protein;PDBTitle: high resolution crystal structure of clpa
68 c2dhrC_
not modelled
98.7
16
PDB header: hydrolaseChain: C: PDB Molecule: ftsh;PDBTitle: whole cytosolic region of atp-dependent metalloprotease2 ftsh (g399l)
69 c1kgsA_
not modelled
98.7
21
PDB header: dna binding proteinChain: A: PDB Molecule: dna binding response regulator d;PDBTitle: crystal structure at 1.50 a of an ompr/phob homolog from thermotoga2 maritima
70 d1jbka_
not modelled
98.7
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain71 c2z4rB_
not modelled
98.7
13
PDB header: dna binding proteinChain: B: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: crystal structure of domain iii from the thermotoga2 maritima replication initiation protein dnaa
72 c3hu2C_
not modelled
98.7
15
PDB header: transport proteinChain: C: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: structure of p97 n-d1 r86a mutant in complex with atpgs
73 d1ixza_
not modelled
98.6
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain74 c3h4mC_
not modelled
98.6
17
PDB header: hydrolaseChain: C: PDB Molecule: proteasome-activating nucleotidase;PDBTitle: aaa atpase domain of the proteasome- activating nucleotidase
75 c1jr3E_
not modelled
98.6
11
PDB header: transferaseChain: E: PDB Molecule: dna polymerase iii, delta' subunit;PDBTitle: crystal structure of the processivity clamp loader gamma2 complex of e. coli dna polymerase iii
76 d2ce7a2
not modelled
98.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain77 c1iy2A_
not modelled
98.6
16
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent metalloprotease ftsh;PDBTitle: crystal structure of the ftsh atpase domain from thermus2 thermophilus
78 c2qz4A_
not modelled
98.5
17
PDB header: hydrolaseChain: A: PDB Molecule: paraplegin;PDBTitle: human paraplegin, aaa domain in complex with adp
79 c1qvrB_
not modelled
98.5
25
PDB header: chaperoneChain: B: PDB Molecule: clpb protein;PDBTitle: crystal structure analysis of clpb
80 d1r7ra3
not modelled
98.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain81 c2p65A_
not modelled
98.5
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pf08_0063;PDBTitle: crystal structure of the first nucleotide binding domain of2 chaperone clpb1, putative, (pv089580) from plasmodium vivax
82 c3pxgA_
not modelled
98.5
12
PDB header: protein bindingChain: A: PDB Molecule: negative regulator of genetic competence clpc/mecb;PDBTitle: structure of meca121 and clpc1-485 complex
83 c2c9oC_
not modelled
98.5
11
PDB header: hydrolaseChain: C: PDB Molecule: ruvb-like 1;PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
84 c3eihB_
not modelled
98.5
15
PDB header: protein transportChain: B: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: crystal structure of s.cerevisiae vps4 in the presence of atpgammas
85 d1l8qa2
not modelled
98.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain86 c2r65A_
not modelled
98.5
16
PDB header: hydrolaseChain: A: PDB Molecule: cell division protease ftsh homolog;PDBTitle: crystal structure of helicobacter pylori atp dependent protease, ftsh2 adp complex
87 c3cf1C_
not modelled
98.4
15
PDB header: transport proteinChain: C: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: structure of p97/vcp in complex with adp/adp.alfx
88 d1ttya_
not modelled
98.4
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain89 c2kjqA_
not modelled
98.4
17
PDB header: replicationChain: A: PDB Molecule: dnaa-related protein;PDBTitle: solution structure of protein nmb1076 from neisseria meningitidis.2 northeast structural genomics consortium target mr101b.
90 c3q9sA_
not modelled
98.4
28
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding response regulator;PDBTitle: crystal structure of rra(1-215) from deinococcus radiodurans
91 d1e32a2
not modelled
98.4
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain92 c1s3sA_
not modelled
98.4
14
PDB header: protein bindingChain: A: PDB Molecule: transitional endoplasmic reticulum atpase (terPDBTitle: crystal structure of aaa atpase p97/vcp nd1 in complex with2 p47 c
93 c3gw4B_
not modelled
98.4
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein from deinococcus2 radiodurans. northeast structural genomics consortium target drr162b.
94 d1smyf2
not modelled
98.3
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain95 d2p7vb1
not modelled
98.3
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain96 d1r6bx3
not modelled
98.3
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain97 c3nbxX_
not modelled
98.2
14
PDB header: hydrolaseChain: X: PDB Molecule: atpase rava;PDBTitle: crystal structure of e. coli rava (regulatory atpase variant a) in2 complex with adp
98 d1rp3a2
not modelled
98.2
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain99 c3hteC_
not modelled
98.2
17
PDB header: motor proteinChain: C: PDB Molecule: atp-dependent clp protease atp-binding subunit clpx;PDBTitle: crystal structure of nucleotide-free hexameric clpx
100 c2x8aA_
not modelled
98.2
13
PDB header: nuclear proteinChain: A: PDB Molecule: nuclear valosin-containing protein-like;PDBTitle: human nuclear valosin containing protein like (nvl), c-2 terminal aaa-atpase domain
101 c3hugA_
not modelled
98.2
30
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
102 d1or7a1
not modelled
98.2
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain103 c2o8xA_
not modelled
98.1
22
PDB header: transcriptionChain: A: PDB Molecule: probable rna polymerase sigma-c factor;PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
104 c1ny5A_
not modelled
98.1
13
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigm54 activator (aaa+ atpase) in the inactive2 state
105 d1xsva_
not modelled
98.1
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like106 d1ku7a_
not modelled
98.1
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain107 d1d2na_
not modelled
98.1
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain108 c2hcbC_
not modelled
98.1
11
PDB header: replicationChain: C: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: structure of amppcp-bound dnaa from aquifex aeolicus
109 d1ny5a2
not modelled
98.1
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain110 d1lv7a_
not modelled
98.1
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain111 c3ulqA_
not modelled
98.1
9
PDB header: gene regulation/transcription activatorChain: A: PDB Molecule: response regulator aspartate phosphatase f;PDBTitle: crystal structure of the anti-activator rapf complexed with the2 response regulator coma dna binding domain
112 c2ifuA_
not modelled
98.0
8
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: gamma-snap;PDBTitle: crystal structure of a gamma-snap from danio rerio
113 c1nsfA_
not modelled
98.0
15
PDB header: protein transportChain: A: PDB Molecule: n-ethylmaleimide sensitive factor;PDBTitle: d2 hexamerization domain of n-ethylmaleimide sensitive factor (nsf)
114 d1g41a_
not modelled
98.0
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain115 d1ku3a_
not modelled
98.0
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain116 c1rp3G_
not modelled
98.0
26
PDB header: transcriptionChain: G: PDB Molecule: rna polymerase sigma factor sigma-28 (flia);PDBTitle: cocrystal structure of the flagellar sigma/anti-sigma2 complex, sigma-28/flgm
117 c3mzyA_
not modelled
98.0
28
PDB header: rna binding proteinChain: A: PDB Molecule: rna polymerase sigma-h factor;PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
118 c3sf4B_
not modelled
98.0
11
PDB header: signaling protein/protein bindingChain: B: PDB Molecule: g-protein-signaling modulator 2;PDBTitle: crystal structure of the complex between the conserved cell polarity2 proteins inscuteable and lgn
119 c1ojlF_
not modelled
97.9
15
PDB header: response regulatorChain: F: PDB Molecule: transcriptional regulatory protein zrar;PDBTitle: crystal structure of a sigma54-activator suggests the2 mechanism for the conformational switch necessary for3 sigma54 binding
120 d1s7oa_
not modelled
97.9
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like