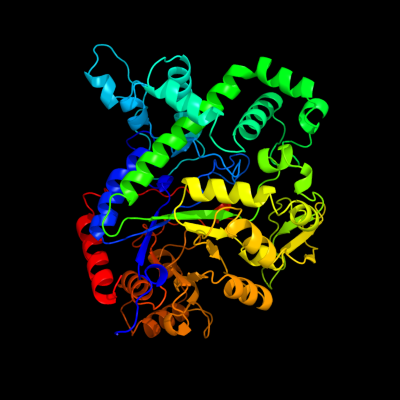
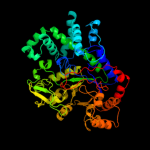
1 d1x1na1
100.0
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain2 d1eswa_
100.0
25
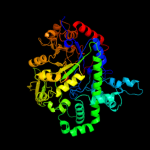
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain3 d1tz7a1
100.0
26
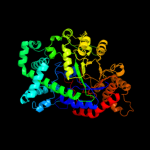
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain4 c3zt5D_
98.6
19
PDB header: hydrolaseChain: D: PDB Molecule: putative glucanohydrolase pep1a;PDBTitle: glge isoform 1 from streptomyces coelicolor with maltose2 bound
5 c3amkA_
98.4
15
PDB header: transferaseChain: A: PDB Molecule: os06g0726400 protein;PDBTitle: structure of the starch branching enzyme i (bei) from oryza sativa l
6 c3amlA_
98.3
17
PDB header: transferaseChain: A: PDB Molecule: os06g0726400 protein;PDBTitle: structure of the starch branching enzyme i (bei) from oryza sativa l
7 c1m53A_
98.2
18
PDB header: isomeraseChain: A: PDB Molecule: isomaltulose synthase;PDBTitle: crystal structure of isomaltulose synthase (pali) from2 klebsiella sp. lx3
8 d1uoka2
98.2
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain9 d1j0ha3
98.2
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain10 c1uokA_
98.1
21
PDB header: glucosidaseChain: A: PDB Molecule: oligo-1,6-glucosidase;PDBTitle: crystal structure of b. cereus oligo-1,6-glucosidase
11 d1bf2a3
98.1
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain12 c1zjaB_
98.1
22
PDB header: isomeraseChain: B: PDB Molecule: trehalulose synthase;PDBTitle: crystal structure of the trehalulose synthase mutb from2 pseudomonas mesoacidophila mx-45 (triclinic form)
13 d1m53a2
98.0
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain14 d1ua7a2
98.0
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain15 d1gvia3
97.9
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain16 c1wpcA_
97.9
19
PDB header: hydrolaseChain: A: PDB Molecule: glucan 1,4-alpha-maltohexaosidase;PDBTitle: crystal structure of maltohexaose-producing amylase complexed with2 pseudo-maltononaose
17 c3a47A_
97.9
17
PDB header: hydrolaseChain: A: PDB Molecule: oligo-1,6-glucosidase;PDBTitle: crystal structure of isomaltase from saccharomyces cerevisiae
18 c1tcmB_
97.9
21
PDB header: glycosyltransferaseChain: B: PDB Molecule: cyclodextrin glycosyltransferase;PDBTitle: cyclodextrin glycosyltransferase w616a mutant from bacillus2 circulans strain 251
19 d1m7xa3
97.9
21
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain20 d1lwha2
97.9
21
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain21 c3dhuC_
not modelled
97.9
26
PDB header: hydrolaseChain: C: PDB Molecule: alpha-amylase;PDBTitle: crystal structure of an alpha-amylase from lactobacillus2 plantarum
22 c1jibA_
not modelled
97.8
16
PDB header: hydrolaseChain: A: PDB Molecule: neopullulanase;PDBTitle: complex of alpha-amylase ii (tva ii) from thermoactinomyces2 vulgaris r-47 with maltotetraose based on a crystal soaked3 with maltohexaose.
23 d2d3na2
not modelled
97.8
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain24 c2ya1A_
not modelled
97.8
24
PDB header: hydrolaseChain: A: PDB Molecule: putative alkaline amylopullulanase;PDBTitle: product complex of a multi-modular glycogen-degrading2 pneumococcal virulence factor spua
25 d2guya2
not modelled
97.8
26
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain26 c2ze0A_
not modelled
97.7
19
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: alpha-glucosidase gsj
27 c2ya0A_
not modelled
97.7
19
PDB header: hydrolaseChain: A: PDB Molecule: putative alkaline amylopullulanase;PDBTitle: catalytic module of the multi-modular glycogen-degrading2 pneumococcal virulence factor spua
28 c2wcsA_
not modelled
97.7
22
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase, catalytic region;PDBTitle: crystal structure of debranching enzyme from nostoc2 punctiforme (npde)
29 c3bmwA_
not modelled
97.7
19
PDB header: transferaseChain: A: PDB Molecule: cyclomaltodextrin glucanotransferase;PDBTitle: cyclodextrin glycosyl transferase from thermoanerobacterium2 thermosulfurigenes em1 mutant s77p complexed with a maltoheptaose3 inhibitor
30 c3k1dA_
not modelled
97.6
19
PDB header: transferaseChain: A: PDB Molecule: 1,4-alpha-glucan-branching enzyme;PDBTitle: crystal structure of glycogen branching enzyme synonym: 1,4-alpha-d-2 glucan:1,4-alpha-d-glucan 6-glucosyl-transferase from mycobacterium3 tuberculosis h37rv
31 c2dh3A_
not modelled
97.6
23
PDB header: transport protein, signaling proteinChain: A: PDB Molecule: 4f2 cell-surface antigen heavy chain;PDBTitle: crystal structure of human ed-4f2hc
32 c1ehaA_
not modelled
97.6
20
PDB header: hydrolaseChain: A: PDB Molecule: glycosyltrehalose trehalohydrolase;PDBTitle: crystal structure of glycosyltrehalose trehalohydrolase2 from sulfolobus solfataricus
33 c1ea9D_
not modelled
97.5
17
PDB header: hydrolaseChain: D: PDB Molecule: cyclomaltodextrinase;PDBTitle: cyclomaltodextrinase
34 d1h3ga3
not modelled
97.4
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain35 c2zidA_
not modelled
97.4
22
PDB header: hydrolaseChain: A: PDB Molecule: dextran glucosidase;PDBTitle: crystal structure of dextran glucosidase e236q complex with2 isomaltotriose
36 d1eh9a3
not modelled
97.3
24
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain37 c2by0A_
not modelled
97.3
21
PDB header: hydrolaseChain: A: PDB Molecule: maltooligosyltrehalose trehalohydrolase;PDBTitle: is radiation damage dependent on the dose-rate used during2 macromolecular crystallography data collection
38 d3bmva4
not modelled
97.2
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain39 d2bhua3
not modelled
97.2
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain40 c2taaA_
not modelled
97.2
28
PDB header: hydrolase (o-glycosyl)Chain: A: PDB Molecule: taka-amylase a;PDBTitle: structure and possible catalytic residues of taka-amylase a
41 c2wskA_
not modelled
97.2
21
PDB header: hydrolaseChain: A: PDB Molecule: glycogen debranching enzyme;PDBTitle: crystal structure of glycogen debranching enzyme glgx from2 escherichia coli k-12
42 c2qpuB_
not modelled
97.2
16
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amylase type a isozyme;PDBTitle: sugar tongs mutant s378p in complex with acarbose
43 d1gcya2
not modelled
97.2
25
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain44 c1hvxA_
not modelled
97.0
15
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: bacillus stearothermophilus alpha-amylase
45 c2z1kA_
not modelled
97.0
23
PDB header: hydrolaseChain: A: PDB Molecule: (neo)pullulanase;PDBTitle: crystal structure of ttha1563 from thermus thermophilus hb8
46 c1gviA_
not modelled
97.0
12
PDB header: hydrolaseChain: A: PDB Molecule: maltogenic amylase;PDBTitle: thermus maltogenic amylase in complex with beta-cd
47 c1qhoA_
not modelled
96.9
16
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: five-domain alpha-amylase from bacillus stearothermophilus,2 maltose/acarbose complex
48 c2wanA_
not modelled
96.8
13
PDB header: hydrolaseChain: A: PDB Molecule: pullulanase;PDBTitle: pullulanase from bacillus acidopullulyticus
49 d1cyga4
not modelled
96.8
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain50 d1ob0a2
not modelled
96.5
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain51 c1cygA_
not modelled
96.4
15
PDB header: glycosyltransferaseChain: A: PDB Molecule: cyclodextrin glucanotransferase;PDBTitle: cyclodextrin glucanotransferase (e.c.2.4.1.19) (cgtase)
52 d1e43a2
not modelled
96.3
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain53 d1pama4
not modelled
96.2
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain54 c1jaeA_
not modelled
96.1
16
PDB header: glycosidaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: structure of tenebrio molitor larval alpha-amylase
55 c2fhfA_
not modelled
96.0
19
PDB header: hydrolaseChain: A: PDB Molecule: pullulanase;PDBTitle: crystal structure analysis of klebsiella pneumoniae2 pullulanase complexed with maltotetraose
56 c3edeB_
not modelled
95.8
24
PDB header: hydrolaseChain: B: PDB Molecule: cyclomaltodextrinase;PDBTitle: structural base for cyclodextrin hydrolysis
57 c1lwhA_
not modelled
95.7
21
PDB header: transferaseChain: A: PDB Molecule: 4-alpha-glucanotransferase;PDBTitle: crystal structure of t. maritima 4-alpha-glucanotransferase
58 d1ht6a2
not modelled
95.7
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain59 d1avaa2
not modelled
95.6
22
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain60 d1ea9c3
not modelled
95.5
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain61 c2gdvA_
not modelled
95.4
10
PDB header: transferaseChain: A: PDB Molecule: sucrose phosphorylase;PDBTitle: sucrose phosphorylase from bifidobacterium adolescentis2 reacted with sucrose
62 c1bplA_
not modelled
95.3
13
PDB header: glycosyltransferaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: glycosyltransferase
63 d1gjwa2
not modelled
95.2
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain64 d2gjpa2
not modelled
95.2
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain65 d1cxla4
not modelled
95.2
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain66 c1gcyA_
not modelled
95.1
14
PDB header: hydrolaseChain: A: PDB Molecule: glucan 1,4-alpha-maltotetrahydrolase;PDBTitle: high resolution crystal structure of maltotetraose-forming2 exo-amylase
67 d2fhfa5
not modelled
95.1
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain68 c2e8yA_
not modelled
94.9
13
PDB header: hydrolaseChain: A: PDB Molecule: amyx protein;PDBTitle: crystal structure of pullulanase type i from bacillus subtilis str.2 168
69 d1ud2a2
not modelled
94.5
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain70 c3k8kB_
not modelled
94.5
19
PDB header: membrane proteinChain: B: PDB Molecule: alpha-amylase, susg;PDBTitle: crystal structure of susg
71 c1jdaA_
not modelled
94.3
13
PDB header: hydrolaseChain: A: PDB Molecule: 1,4-alpha maltotetrahydrolase;PDBTitle: maltotetraose-forming exo-amylase
72 d1r7aa2
not modelled
94.3
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain73 d2aaaa2
not modelled
94.0
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain74 c1gjuA_
not modelled
94.0
27
PDB header: transferaseChain: A: PDB Molecule: maltodextrin glycosyltransferase;PDBTitle: maltosyltransferase from thermotoga maritima
75 c1m7xC_
not modelled
94.0
16
PDB header: transferaseChain: C: PDB Molecule: 1,4-alpha-glucan branching enzyme;PDBTitle: the x-ray crystallographic structure of branching enzyme
76 c3faxA_
not modelled
93.7
24
PDB header: hydrolaseChain: A: PDB Molecule: reticulocyte binding protein;PDBTitle: the crystal structure of gbs pullulanase sap in complex with2 maltotetraose
77 c1jgiA_
not modelled
93.4
22
PDB header: transferaseChain: A: PDB Molecule: amylosucrase;PDBTitle: crystal structure of the active site mutant glu328gln of2 amylosucrase from neisseria polysaccharea in complex with3 the natural substrate sucrose
78 c3bc9A_
not modelled
93.3
18
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase, catalytic region;PDBTitle: alpha-amylase b in complex with acarbose
79 c1e40A_
not modelled
93.3
18
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: tris/maltotriose complex of chimaeric amylase from b.2 amyloliquefaciens and b. licheniformis at 2.2a
80 c2x4bA_
not modelled
93.0
14
PDB header: hydrolaseChain: A: PDB Molecule: limit dextrinase;PDBTitle: barley limit dextrinase in complex with beta-cyclodextrin
81 c1bf2A_
not modelled
92.9
15
PDB header: hydrolaseChain: A: PDB Molecule: isoamylase;PDBTitle: structure of pseudomonas isoamylase
82 c3czkA_
not modelled
92.6
8
PDB header: hydrolaseChain: A: PDB Molecule: sucrose hydrolase;PDBTitle: crystal structure analysis of sucrose hydrolase(suh) e322q-2 sucrose complex
83 d1mxga2
not modelled
92.5
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain84 c2aaaA_
not modelled
92.5
17
PDB header: glycosidaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: calcium binding in alpha-amylases: an x-ray diffraction2 study at 2.1 angstroms resolution of two enzymes from3 aspergillus
85 c1jd7A_
not modelled
92.3
13
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: crystal structure analysis of the mutant k300r of2 pseudoalteromonas haloplanctis alpha-amylase
86 c3m07A_
not modelled
92.3
19
PDB header: unknown functionChain: A: PDB Molecule: putative alpha amylase;PDBTitle: 1.4 angstrom resolution crystal structure of putative alpha2 amylase from salmonella typhimurium.
87 c3blpX_
not modelled
91.9
18
PDB header: hydrolaseChain: X: PDB Molecule: alpha-amylase 1;PDBTitle: role of aromatic residues in human salivary alpha-amylase
88 d1hvxa2
not modelled
91.8
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain89 d1g5aa2
not modelled
91.5
28
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain90 c1bplB_
not modelled
91.5
12
PDB header: glycosyltransferaseChain: B: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: glycosyltransferase
91 d1qhoa4
not modelled
91.3
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain92 d1wzla3
not modelled
90.8
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain93 d1hx0a2
not modelled
90.5
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain94 c1bagA_
not modelled
90.2
23
PDB header: alpha-amylaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: alpha-amylase from bacillus subtilis complexed with2 maltopentaose
95 d1jaea2
not modelled
89.5
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain96 c1wzaA_
not modelled
88.7
23
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase a;PDBTitle: crystal structure of alpha-amylase from h.orenii
97 d1g94a2
not modelled
88.6
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain98 d3dhpa2
not modelled
87.9
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain99 c2yfnA_
not modelled
87.2
11
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase-sucrose kinase agask;PDBTitle: galactosidase domain of alpha-galactosidase-sucrose kinase,2 agask
100 c2vncB_
not modelled
86.6
19
PDB header: hydrolaseChain: B: PDB Molecule: glycogen operon protein glgx;PDBTitle: crystal structure of glycogen debranching enzyme trex from2 sulfolobus solfataricus
101 d1ji1a3
not modelled
86.3
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain102 d1wzaa2
not modelled
83.4
25
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain103 d1cgta4
not modelled
82.9
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain104 c1mwoA_
not modelled
80.0
16
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase;PDBTitle: crystal structure analysis of the hyperthermostable2 pyrocoocus woesei alpha-amylase
105 c1ud8A_
not modelled
79.2
14
PDB header: hydrolaseChain: A: PDB Molecule: amylase;PDBTitle: crystal structure of amyk38 with lithium ion
106 c2bnoA_
75.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: epoxidase;PDBTitle: the structure of hydroxypropylphosphonic acid epoxidase2 from s. wedmorenis.
107 c2d0gA_
not modelled
73.5
20
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase i;PDBTitle: crystal structure of thermoactinomyces vulgaris r-47 alpha-2 amylase 1 (tvai) mutant d356n/e396q complexed with p5, a3 pullulan model oligosaccharide
108 c3hjeA_
not modelled
73.4
17
PDB header: transferaseChain: A: PDB Molecule: 704aa long hypothetical glycosyltransferase;PDBTitle: crystal structure of sulfolobus tokodaii hypothetical2 maltooligosyl trehalose synthase
109 c3cc1B_
not modelled
65.1
12
PDB header: hydrolaseChain: B: PDB Molecule: putative alpha-n-acetylgalactosaminidase;PDBTitle: crystal structure of a putative alpha-n-acetylgalactosaminidase2 (bh1870) from bacillus halodurans c-125 at 2.00 a resolution
110 d1j0ha2
63.2
21
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain111 d1h9aa1
not modelled
62.8
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain112 c2bhlB_
not modelled
61.4
20
PDB header: oxidoreductase (choh(d)-nadp)Chain: B: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase2 (deletion variant) complexed with glucose-6-phosphate
113 d2a6ca1
not modelled
60.9
17
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE1354114 c1qkiE_
not modelled
59.7
20
PDB header: oxidoreductaseChain: E: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase2 (variant canton r459l) complexed with structural nadp+
115 c3pxpA_
not modelled
58.7
20
PDB header: transcription regulatorChain: A: PDB Molecule: helix-turn-helix domain protein;PDBTitle: crystal structure of a pas and dna binding domain containing protein2 (caur_2278) from chloroflexus aurantiacus j-10-fl at 2.30 a3 resolution
116 d1l1ya_
not modelled
58.7
25
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain117 c1l2aD_
not modelled
58.7
25
PDB header: hydrolaseChain: D: PDB Molecule: cellobiohydrolase;PDBTitle: the crystal structure and catalytic mechanism of2 cellobiohydrolase cels, the major enzymatic component of3 the clostridium thermocellum cellulosome
118 d2ofya1
not modelled
56.9
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like119 d1g9ga_
not modelled
55.9
21
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain120 c3gzaB_
not modelled
55.6
16
PDB header: hydrolaseChain: B: PDB Molecule: putative alpha-l-fucosidase;PDBTitle: crystal structure of putative alpha-l-fucosidase (np_812709.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.60 a resolution