| 1 | d1tv8a_ |
|
 |
100.0 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:MoCo biosynthesis proteins |
| 2 | c2yx0A_ |
|
 |
99.9 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
| 3 | c3c8fA_ |
|
 |
99.8 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate formate-lyase 1-activating enzyme;
PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
|
| 4 | c1r30A_ |
|
 |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
|
| 5 | d1r30a_ |
|
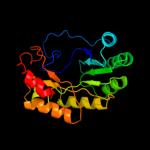 |
99.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
| 6 | c3t7vA_ |
|
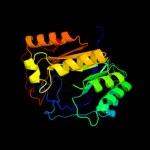 |
99.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
| 7 | c2a5hC_ |
|
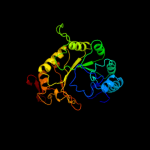 |
99.8 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
| 8 | c3cixA_ |
|
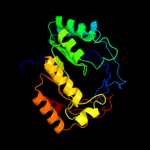 |
99.7 |
14 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
| 9 | c3rfaA_ |
|
 |
99.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
| 10 | c2z2uA_ |
|
 |
99.5 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
| 11 | d1olta_ |
|
 |
99.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
| 12 | c3canA_ |
|
 |
99.5 |
17 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
| 13 | c2qgqF_ |
|
 |
99.1 |
9 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:protein tm_1862;
PDBTitle: crystal structure of tm_1862 from thermotoga maritima.2 northeast structural genomics consortium target vr77
|
| 14 | c3ivuB_ |
|
 |
97.4 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 15 | c3eegB_ |
|
 |
95.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 16 | c3ewbX_ |
|
 |
95.0 |
9 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
|
| 17 | c2ftpA_ |
|
 |
92.7 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
| 18 | c1nvmG_ |
|
 |
91.6 |
15 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 19 | c2cw6B_ |
|
 |
90.9 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
| 20 | c3bleA_ |
|
 |
90.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 21 | d1nvma2 |
|
not modelled |
89.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 22 | c1ydnA_ |
|
not modelled |
89.5 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 23 | c1ydoC_ |
|
not modelled |
87.8 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
| 24 | c1sr9A_ |
|
not modelled |
82.2 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of leua from mycobacterium tuberculosis
|
| 25 | c2nx9B_ |
|
not modelled |
82.1 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
| 26 | d1qt1a_ |
|
not modelled |
81.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 27 | c3khdC_ |
|
not modelled |
79.4 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pff1300w.
|
| 28 | c2wcsA_ |
|
not modelled |
75.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha amylase, catalytic region;
PDBTitle: crystal structure of debranching enzyme from nostoc2 punctiforme (npde)
|
| 29 | d1qhoa4 |
|
not modelled |
74.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 30 | c2zyfA_ |
|
not modelled |
70.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:homocitrate synthase;
PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with magnesuim ion and alpha-ketoglutarate
|
| 31 | d2aaaa2 |
|
not modelled |
70.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 32 | d1muwa_ |
|
not modelled |
68.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 33 | d3bmva4 |
|
not modelled |
68.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 34 | c1lwhA_ |
|
not modelled |
67.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:4-alpha-glucanotransferase;
PDBTitle: crystal structure of t. maritima 4-alpha-glucanotransferase
|
| 35 | d1pama4 |
|
not modelled |
66.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 36 | c1m53A_ |
|
not modelled |
66.5 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:isomaltulose synthase;
PDBTitle: crystal structure of isomaltulose synthase (pali) from2 klebsiella sp. lx3
|
| 37 | c3hpxB_ |
|
not modelled |
65.3 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of mycobacterium tuberculosis leua active site2 domain 1-425 (truncation mutant delta:426-644)
|
| 38 | c3a47A_ |
|
not modelled |
64.6 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:oligo-1,6-glucosidase;
PDBTitle: crystal structure of isomaltase from saccharomyces cerevisiae
|
| 39 | d1cgta4 |
|
not modelled |
64.0 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 40 | d1bf2a3 |
|
not modelled |
64.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 41 | d1h3ga3 |
|
not modelled |
63.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 42 | d1gjwa2 |
|
not modelled |
63.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 43 | d1uoka2 |
|
not modelled |
61.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 44 | d1cxla4 |
|
not modelled |
58.4 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 45 | c1ea9D_ |
|
not modelled |
58.0 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:cyclomaltodextrinase;
PDBTitle: cyclomaltodextrinase
|
| 46 | d1m53a2 |
|
not modelled |
57.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 47 | c2aaaA_ |
|
not modelled |
56.5 |
10 |
PDB header:glycosidase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: calcium binding in alpha-amylases: an x-ray diffraction2 study at 2.1 angstroms resolution of two enzymes from3 aspergillus
|
| 48 | d1gvia3 |
|
not modelled |
56.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 49 | c1uokA_ |
|
not modelled |
55.7 |
16 |
PDB header:glucosidase
Chain: A: PDB Molecule:oligo-1,6-glucosidase;
PDBTitle: crystal structure of b. cereus oligo-1,6-glucosidase
|
| 50 | d2guya2 |
|
not modelled |
54.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 51 | d1e43a2 |
|
not modelled |
53.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 52 | c1qhoA_ |
|
not modelled |
52.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: five-domain alpha-amylase from bacillus stearothermophilus,2 maltose/acarbose complex
|
| 53 | d1g5aa2 |
|
not modelled |
52.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 54 | d1lwha2 |
|
not modelled |
50.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 55 | c2ze0A_ |
|
not modelled |
50.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-glucosidase;
PDBTitle: alpha-glucosidase gsj
|
| 56 | c2zidA_ |
|
not modelled |
49.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:dextran glucosidase;
PDBTitle: crystal structure of dextran glucosidase e236q complex with2 isomaltotriose
|
| 57 | c3edeB_ |
|
not modelled |
49.7 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:cyclomaltodextrinase;
PDBTitle: structural base for cyclodextrin hydrolysis
|
| 58 | c1bf2A_ |
|
not modelled |
48.9 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:isoamylase;
PDBTitle: structure of pseudomonas isoamylase
|
| 59 | c2ya0A_ |
|
not modelled |
48.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alkaline amylopullulanase;
PDBTitle: catalytic module of the multi-modular glycogen-degrading2 pneumococcal virulence factor spua
|
| 60 | d1ob0a2 |
|
not modelled |
48.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 61 | c3dxiB_ |
|
not modelled |
48.6 |
9 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of the n-terminal domain of a putative2 aldolase (bvu_2661) from bacteroides vulgatus
|
| 62 | c1gjuA_ |
|
not modelled |
48.4 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:maltodextrin glycosyltransferase;
PDBTitle: maltosyltransferase from thermotoga maritima
|
| 63 | c2qpuB_ |
|
not modelled |
47.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-amylase type a isozyme;
PDBTitle: sugar tongs mutant s378p in complex with acarbose
|
| 64 | d1ea9c3 |
|
not modelled |
47.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 65 | c1jibA_ |
|
not modelled |
46.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:neopullulanase;
PDBTitle: complex of alpha-amylase ii (tva ii) from thermoactinomyces2 vulgaris r-47 with maltotetraose based on a crystal soaked3 with maltohexaose.
|
| 66 | c2z1kA_ |
|
not modelled |
46.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:(neo)pullulanase;
PDBTitle: crystal structure of ttha1563 from thermus thermophilus hb8
|
| 67 | d1yhta1 |
|
not modelled |
45.8 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 68 | c2zq0B_ |
|
not modelled |
45.7 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-glucosidase (alpha-glucosidase susb);
PDBTitle: crystal structure of susb complexed with acarbose
|
| 69 | d2gjpa2 |
|
not modelled |
44.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 70 | c2jvfA_ |
|
not modelled |
44.1 |
12 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo protein m7;
PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
|
| 71 | c3czkA_ |
|
not modelled |
43.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:sucrose hydrolase;
PDBTitle: crystal structure analysis of sucrose hydrolase(suh) e322q-2 sucrose complex
|
| 72 | d1u7pa_ |
|
not modelled |
42.9 |
8 |
Fold:HAD-like
Superfamily:HAD-like
Family:Magnesium-dependent phosphatase-1, Mdp1 |
| 73 | c1zjaB_ |
|
not modelled |
42.8 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:trehalulose synthase;
PDBTitle: crystal structure of the trehalulose synthase mutb from2 pseudomonas mesoacidophila mx-45 (triclinic form)
|
| 74 | c1qysA_ |
|
not modelled |
42.3 |
9 |
PDB header:de novo protein
Chain: A: PDB Molecule:top7;
PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
|
| 75 | d1ua7a2 |
|
not modelled |
42.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 76 | c3dhuC_ |
|
not modelled |
41.9 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure of an alpha-amylase from lactobacillus2 plantarum
|
| 77 | c1ehaA_ |
|
not modelled |
41.5 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyltrehalose trehalohydrolase;
PDBTitle: crystal structure of glycosyltrehalose trehalohydrolase2 from sulfolobus solfataricus
|
| 78 | c1gviA_ |
|
not modelled |
40.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:maltogenic amylase;
PDBTitle: thermus maltogenic amylase in complex with beta-cd
|
| 79 | d1j0ha3 |
|
not modelled |
40.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 80 | c2taaA_ |
|
not modelled |
39.2 |
11 |
PDB header:hydrolase (o-glycosyl)
Chain: A: PDB Molecule:taka-amylase a;
PDBTitle: structure and possible catalytic residues of taka-amylase a
|
| 81 | d1cyga4 |
|
not modelled |
39.2 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 82 | c2dh3A_ |
|
not modelled |
38.1 |
11 |
PDB header:transport protein, signaling protein
Chain: A: PDB Molecule:4f2 cell-surface antigen heavy chain;
PDBTitle: crystal structure of human ed-4f2hc
|
| 83 | c3k8kB_ |
|
not modelled |
38.1 |
19 |
PDB header:membrane protein
Chain: B: PDB Molecule:alpha-amylase, susg;
PDBTitle: crystal structure of susg
|
| 84 | d1ud2a2 |
|
not modelled |
37.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 85 | c1bagA_ |
|
not modelled |
36.7 |
17 |
PDB header:alpha-amylase
Chain: A: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: alpha-amylase from bacillus subtilis complexed with2 maltopentaose
|
| 86 | c1jgiA_ |
|
not modelled |
36.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:amylosucrase;
PDBTitle: crystal structure of the active site mutant glu328gln of2 amylosucrase from neisseria polysaccharea in complex with3 the natural substrate sucrose
|
| 87 | c1cygA_ |
|
not modelled |
35.9 |
10 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:cyclodextrin glucanotransferase;
PDBTitle: cyclodextrin glucanotransferase (e.c.2.4.1.19) (cgtase)
|
| 88 | c3gh7A_ |
|
not modelled |
35.7 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of beta-hexosaminidase from paenibacillus2 sp. ts12 in complex with galnac
|
| 89 | d1jaka1 |
|
not modelled |
34.8 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 90 | d1mzha_ |
|
not modelled |
34.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 91 | d1z6ma1 |
|
not modelled |
33.8 |
18 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:DsbA-like |
| 92 | c2yl8A_ |
|
not modelled |
33.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
| 93 | d1ht6a2 |
|
not modelled |
33.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 94 | c3os4A_ |
|
not modelled |
32.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: the crystal structure of nicotinate phosphoribosyltransferase from2 yersinia pestis
|
| 95 | c3na8A_ |
|
not modelled |
32.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 96 | c2rfgB_ |
|
not modelled |
31.8 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 97 | d1pkla2 |
|
not modelled |
31.7 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 98 | c3bmwA_ |
|
not modelled |
31.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:cyclomaltodextrin glucanotransferase;
PDBTitle: cyclodextrin glycosyl transferase from thermoanerobacterium2 thermosulfurigenes em1 mutant s77p complexed with a maltoheptaose3 inhibitor
|
| 99 | c3zt5D_ |
|
not modelled |
31.4 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative glucanohydrolase pep1a;
PDBTitle: glge isoform 1 from streptomyces coelicolor with maltose2 bound
|
| 100 | d1yx1a1 |
|
not modelled |
30.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
| 101 | c3ngjC_ |
|
not modelled |
30.0 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 entamoeba histolytica
|
| 102 | c2ya1A_ |
|
not modelled |
29.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alkaline amylopullulanase;
PDBTitle: product complex of a multi-modular glycogen-degrading2 pneumococcal virulence factor spua
|
| 103 | d1o0ya_ |
|
not modelled |
29.5 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 104 | d1mxga2 |
|
not modelled |
28.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 105 | d1avaa2 |
|
not modelled |
27.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 106 | c1bplA_ |
|
not modelled |
27.3 |
13 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: glycosyltransferase
|
| 107 | d1wzla3 |
|
not modelled |
27.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 108 | c3faxA_ |
|
not modelled |
27.0 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:reticulocyte binding protein;
PDBTitle: the crystal structure of gbs pullulanase sap in complex with2 maltotetraose
|
| 109 | d1yira1 |
|
not modelled |
26.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase C-terminal domain |
| 110 | c2kl5A_ |
|
not modelled |
25.9 |
55 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yutd;
PDBTitle: solution nmr structure of protein yutd from b.subtilis, northeast2 structural genomics consortium target sr232
|
| 111 | d1d3ga_ |
|
not modelled |
25.5 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 112 | d1nowa1 |
|
not modelled |
25.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 113 | d1mioa_ |
|
not modelled |
24.8 |
8 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 114 | c3n2xB_ |
|
not modelled |
24.7 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 115 | c2epoB_ |
|
not modelled |
24.6 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:n-acetyl-beta-d-glucosaminidase;
PDBTitle: n-acetyl-b-d-glucosaminidase (gcna) from streptococcus gordonii
|
| 116 | c1e40A_ |
|
not modelled |
24.3 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: tris/maltotriose complex of chimaeric amylase from b.2 amyloliquefaciens and b. licheniformis at 2.2a
|
| 117 | d1pbya1 |
|
not modelled |
23.6 |
13 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
| 118 | d2bhua3 |
|
not modelled |
23.5 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 119 | c1tcmB_ |
|
not modelled |
23.4 |
10 |
PDB header:glycosyltransferase
Chain: B: PDB Molecule:cyclodextrin glycosyltransferase;
PDBTitle: cyclodextrin glycosyltransferase w616a mutant from bacillus2 circulans strain 251
|
| 120 | c1ud8A_ |
|
not modelled |
22.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:amylase;
PDBTitle: crystal structure of amyk38 with lithium ion
|

