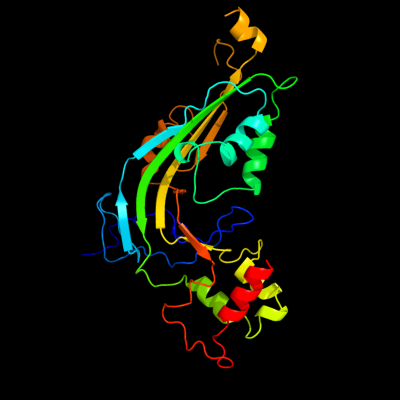
| 1 | c2qqhA_
|
|
 |
100.0 |
20 |
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:complement component c8 alpha chain;
PDBTitle: structure of c8a-macpf reveals mechanism of membrane attack2 in complement immune defense
|
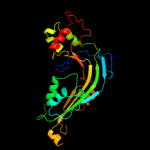
| 2 | c2rd7A_
|
|
 |
100.0 |
20 |
PDB header:immune system
Chain: A: PDB Molecule:complement component c8 alpha chain;
PDBTitle: human complement membrane attack proteins share a common2 fold with bacterial cytolysins
|
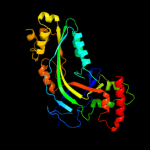
| 3 | c3nsjA_
|
|
 |
100.0 |
21 |
PDB header:immune system
Chain: A: PDB Molecule:perforin-1;
PDBTitle: the x-ray crystal structure of lymphocyte perforin
|
| 4 | c2qp2A_
|
|
 |
99.9 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:unknown protein;
PDBTitle: structure of a macpf/perforin-like protein
|
| 5 | c3kk7B_
|
|
 |
99.2 |
18 |
PDB header:cell invasion
Chain: B: PDB Molecule:putative cell invasion protein with mac/perforin domain;
PDBTitle: crystal structure of putative cell invasion protein with mac/perforin2 domain (np_812351.1) from bacteriodes thetaiotaomicron vpi-5482 at3 2.46 a resolution
|
| 6 | c3ojyA_
|
|
 |
96.4 |
21 |
PDB header:immune system
Chain: A: PDB Molecule:complement component c8 alpha chain;
PDBTitle: crystal structure of human complement component c8
|
| 7 | c3t5oA_
|
|
 |
86.4 |
18 |
PDB header:immune system
Chain: A: PDB Molecule:complement component c6;
PDBTitle: crystal structure of human complement component c6
|
| 8 | c2m20B_
|
|
 |
72.1 |
19 |
PDB header:signaling protein
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: egfr transmembrane - juxtamembrane (tm-jm) segment in bicelles: md2 guided nmr refined structure.
|
| 9 | c2kncB_
|
|
 |
64.2 |
19 |
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 10 | c3ojyB_
|
|
 |
54.6 |
15 |
PDB header:immune system
Chain: B: PDB Molecule:complement component c8 beta chain;
PDBTitle: crystal structure of human complement component c8
|
| 11 | c2kr1A_
|
|
 |
33.4 |
28 |
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin protein ligase e3a;
PDBTitle: solution nmr structure of zinc binding n-terminal domain of ubiquitin-2 protein ligase e3a from homo sapiens. northeast structural genomics3 consortium (nesg) target hr3662
|
| 12 | c2kncA_
|
|
 |
32.6 |
18 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 13 | c1ihmC_
|
|
 |
27.0 |
18 |
PDB header:virus
Chain: C: PDB Molecule:capsid protein;
PDBTitle: crystal structure analysis of norwalk virus capsid
|
| 14 | d1ihma_
|
|
 |
27.0 |
18 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Caliciviridae-like VP |
| 15 | c3floD_
|
|
 |
23.8 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:dna polymerase alpha catalytic subunit a;
PDBTitle: crystal structure of the carboxyl-terminal domain of yeast dna2 polymerase alpha in complex with its b subunit
|
| 16 | c2k9yB_
|
|
 |
23.8 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
|
| 17 | c2k9yA_
|
|
 |
23.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
|
| 18 | c1adyA_
|
|
 |
22.6 |
16 |
PDB header:trna synthetase
Chain: A: PDB Molecule:histidyl-trna synthetase;
PDBTitle: histidyl-trna synthetase in complex with histidyl-adenylate
|
| 19 | c2qbxB_
|
|
 |
21.2 |
29 |
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-b receptor 2;
PDBTitle: ephb2/snew antagonistic peptide complex
|
| 20 | c2ks1B_
|
|
 |
20.5 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
|
| 21 | c2dk6A_ |
|
not modelled |
18.6 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:parp11 protein;
PDBTitle: solution structure of wwe domain in poly (adp-ribose)2 polymerase family, member 11 (parp 11)
|
| 22 | c2auhB_ |
|
not modelled |
17.8 |
55 |
PDB header:transferase/signaling protein
Chain: B: PDB Molecule:growth factor receptor-bound protein 14;
PDBTitle: crystal structure of the grb14 bps region in complex with2 the insulin receptor tyrosine kinase
|
| 23 | d2g38b1 |
|
not modelled |
16.8 |
36 |
Fold:Ferritin-like
Superfamily:PE/PPE dimer-like
Family:PPE |
| 24 | c2g38B_ |
|
not modelled |
16.8 |
36 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:ppe family protein;
PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
|
| 25 | c3p1iC_ |
|
not modelled |
16.2 |
29 |
PDB header:transferase,signaling protein
Chain: C: PDB Molecule:ephrin type-b receptor 3;
PDBTitle: ligand binding domain of human ephrin type-b receptor 3
|
| 26 | c3o14B_ |
|
not modelled |
16.1 |
14 |
PDB header:gene regulation
Chain: B: PDB Molecule:anti-ecfsigma factor, chrr;
PDBTitle: crystal structure of an anti-ecfsigma factor, chrr (maqu_0586) from2 marinobacter aquaeolei vt8 at 1.70 a resolution
|
| 27 | c3gr0D_ |
|
not modelled |
15.9 |
13 |
PDB header:membrane protein
Chain: D: PDB Molecule:protein prgh;
PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-362)
|
| 28 | c3dkbA_ |
|
not modelled |
15.7 |
43 |
PDB header:hydrolase
Chain: A: PDB Molecule:tumor necrosis factor, alpha-induced protein 3;
PDBTitle: crystal structure of a20, 2.5 angstrom
|
| 29 | c2cpbA_ |
|
not modelled |
15.4 |
31 |
PDB header:viral protein
Chain: A: PDB Molecule:m13 major coat protein;
PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
|
| 30 | c2l2tA_ |
|
not modelled |
15.4 |
41 |
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
|
| 31 | c2lcxB_ |
|
not modelled |
15.4 |
41 |
PDB header:transferase
Chain: B: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: spatial structure of the erbb4 dimeric tm domain
|
| 32 | c2nyuA_ |
|
not modelled |
15.3 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative ribosomal rna methyltransferase 2;
PDBTitle: crystal structure of human ftsj homolog 2 (e.coli) protein2 in complex with s-adenosylmethionine
|
| 33 | d1w7ja1 |
|
not modelled |
15.3 |
45 |
Fold:SH3-like barrel
Superfamily:Myosin S1 fragment, N-terminal domain
Family:Myosin S1 fragment, N-terminal domain |
| 34 | c2kr0A_ |
|
not modelled |
15.1 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:proteasomal ubiquitin receptor adrm1;
PDBTitle: a proteasome protein
|
| 35 | c2ko5A_ |
|
not modelled |
14.7 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:ring finger protein z;
PDBTitle: nmr solution structure of lfv-z
|
| 36 | c3gr1A_ |
|
not modelled |
14.2 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:protein prgh;
PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
|
| 37 | d1rp4a_ |
|
not modelled |
13.7 |
17 |
Fold:ERO1-like
Superfamily:ERO1-like
Family:ERO1-like |
| 38 | d1v5ra1 |
|
not modelled |
13.6 |
42 |
Fold:N domain of copper amine oxidase-like
Superfamily:GAS2 domain-like
Family:GAS2 domain |
| 39 | c2lohA_ |
|
not modelled |
13.6 |
19 |
PDB header:neuropeptide
Chain: A: PDB Molecule:p3(42);
PDBTitle: dimeric structure of transmembrane domain of amyloid precursor protein2 in micellar environment
|
| 40 | d1c3ab_ |
|
not modelled |
13.5 |
23 |
Fold:C-type lectin-like
Superfamily:C-type lectin-like
Family:C-type lectin domain |
| 41 | c3h3iA_ |
|
not modelled |
13.1 |
22 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:putative lipid binding protein;
PDBTitle: crystal structure of a putative lipid binding protein (bt_2261) from2 bacteroides thetaiotaomicron vpi-5482 at 2.20 a resolution
|
| 42 | c2l8sA_ |
|
not modelled |
13.0 |
11 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-1;
PDBTitle: solution nmr structure of transmembrane and cytosolic regions of2 integrin alpha1 in detergent micelles
|
| 43 | d1v7pa_ |
|
not modelled |
12.6 |
36 |
Fold:C-type lectin-like
Superfamily:C-type lectin-like
Family:C-type lectin domain |
| 44 | c4bq7D_ |
|
not modelled |
11.6 |
40 |
PDB header:cell adhesion
Chain: D: PDB Molecule:rgm domain family member b;
PDBTitle: crystal structure of the rgmb-neo1 complex form 2
|
| 45 | c1ifpA_ |
|
not modelled |
11.6 |
20 |
PDB header:virus
Chain: A: PDB Molecule:major coat protein assembly;
PDBTitle: inovirus (filamentous bacteriophage) strain pf3 major coat2 protein assembly
|
| 46 | c3douA_ |
|
not modelled |
11.1 |
50 |
PDB header:transferase
Chain: A: PDB Molecule:ribosomal rna large subunit methyltransferase j;
PDBTitle: crystal structure of methyltransferase involved in cell2 division from thermoplasma volcanicum gss1
|
| 47 | c3m8lA_ |
|
not modelled |
10.9 |
19 |
PDB header:virus
Chain: A: PDB Molecule:capsid protein;
PDBTitle: crystal structure analysis of the feline calicivirus capsid protein
|
| 48 | c1kg1A_ |
|
not modelled |
10.8 |
33 |
PDB header:toxin
Chain: A: PDB Molecule:necrosis inducing protein 1;
PDBTitle: nmr structure of the nip1 elicitor protein from2 rhynchosporium secalis
|
| 49 | d1kg1a_ |
|
not modelled |
10.8 |
33 |
Fold:Necrosis inducing protein 1, NIP1
Superfamily:Necrosis inducing protein 1, NIP1
Family:Necrosis inducing protein 1, NIP1 |
| 50 | d1k1xa1 |
|
not modelled |
10.6 |
38 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Families 57/38 glycoside transferase middle domain
Family:4-alpha-glucanotransferase, domain 2 |
| 51 | c2kpeB_ |
|
not modelled |
10.5 |
35 |
PDB header:membrane protein
Chain: B: PDB Molecule:glycophorin-a;
PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
|
| 52 | c2kpeA_ |
|
not modelled |
10.5 |
35 |
PDB header:membrane protein
Chain: A: PDB Molecule:glycophorin-a;
PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
|
| 53 | c1afoB_ |
|
not modelled |
10.2 |
31 |
PDB header:integral membrane protein
Chain: B: PDB Molecule:glycophorin a;
PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
|
| 54 | c2xrfA_ |
|
not modelled |
10.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:uridine phosphorylase 2;
PDBTitle: crystal structure of human uridine phosphorylase 2
|
| 55 | c1h3tB_ |
|
not modelled |
10.1 |
17 |
PDB header:immune system
Chain: B: PDB Molecule:ig gamma-1 chain c region;
PDBTitle: crystal structure of the human igg1 fc-fragment,glycoform2 (mn2f)2
|
| 56 | c4a18P_ |
|
not modelled |
9.8 |
23 |
PDB header:ribosome
Chain: P: PDB Molecule:rpl38;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
|
| 57 | c3k44D_ |
|
not modelled |
9.7 |
33 |
PDB header:nucleic acid binding protein
Chain: D: PDB Molecule:purine-rich binding protein-alpha, isoform b;
PDBTitle: crystal structure of drosophila melanogaster pur-alpha
|
| 58 | d1hdla_ |
|
not modelled |
9.7 |
16 |
Fold:Kazal-type serine protease inhibitors
Superfamily:Kazal-type serine protease inhibitors
Family:Serine proteinase inhibitor lekti |
| 59 | c2e2wA_ |
|
not modelled |
9.6 |
33 |
PDB header:ligase
Chain: A: PDB Molecule:dna ligase 4;
PDBTitle: solution structure of the first brct domain of human dna2 ligase iv
|
| 60 | c4g84B_ |
|
not modelled |
9.6 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:histidine--trna ligase, cytoplasmic;
PDBTitle: crystal structure of human hisrs
|
| 61 | c3mkuA_ |
|
not modelled |
9.6 |
22 |
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
|
| 62 | c4esnB_ |
|
not modelled |
9.4 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a hypothetical protein (rumgna_02503) from2 ruminococcus gnavus atcc 29149 at 2.20 a resolution
|
| 63 | d3bdwa1 |
|
not modelled |
9.3 |
21 |
Fold:C-type lectin-like
Superfamily:C-type lectin-like
Family:C-type lectin domain |
| 64 | c4a53A_ |
|
not modelled |
9.2 |
50 |
PDB header:rna binding protein
Chain: A: PDB Molecule:edc3;
PDBTitle: structural basis of the dcp1:dcp2 mrna decapping complex activation2 by edc3 and scd6
|
| 65 | c4djiA_ |
|
not modelled |
9.2 |
6 |
PDB header:transport protein
Chain: A: PDB Molecule:probable glutamate/gamma-aminobutyrate antiporter;
PDBTitle: structure of glutamate-gaba antiporter gadc
|
| 66 | c4iopB_ |
|
not modelled |
9.1 |
14 |
PDB header:immune system
Chain: B: PDB Molecule:killer cell lectin-like receptor subfamily f member 2;
PDBTitle: crystal structure of nkp65 bound to its ligand kacl
|
| 67 | c3m9zA_ |
|
not modelled |
9.0 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:killer cell lectin-like receptor subfamily b member 1a;
PDBTitle: crystal structure of extracellular domain of mouse nkr-p1a
|
| 68 | c3zrhA_ |
|
not modelled |
9.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:ubiquitin thioesterase zranb1;
PDBTitle: crystal structure of the lys29, lys33-linkage-specific trabid otu2 deubiquitinase domain reveals an ankyrin-repeat ubiquitin binding3 domain (ankubd)
|
| 69 | d1pfoa_ |
|
not modelled |
9.0 |
10 |
Fold:Perfringolysin
Superfamily:Perfringolysin
Family:Perfringolysin |
| 70 | c1pfoA_ |
|
not modelled |
9.0 |
10 |
PDB header:toxin
Chain: A: PDB Molecule:perfringolysin o;
PDBTitle: perfringolysin o
|
| 71 | d1c3aa_ |
|
not modelled |
9.0 |
60 |
Fold:C-type lectin-like
Superfamily:C-type lectin-like
Family:C-type lectin domain |
| 72 | c2k5jB_ |
|
not modelled |
9.0 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein yiif;
PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
|
| 73 | c2k1aA_ |
|
not modelled |
8.9 |
17 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
|
| 74 | d2rgfa_ |
|
not modelled |
8.9 |
22 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ras-binding domain, RBD |
| 75 | c3a9lB_ |
|
not modelled |
8.7 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:poly-gamma-glutamate hydrolase;
PDBTitle: structure of bacteriophage poly-gamma-glutamate hydrolase
|
| 76 | c3zf7p_ |
|
not modelled |
8.7 |
14 |
PDB header:ribosome
Chain: P: PDB Molecule:probable 60s ribosomal protein l14;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
| 77 | d2zkmx4 |
|
not modelled |
8.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Mammalian PLC |
| 78 | c1b5fC_ |
|
not modelled |
8.6 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein (cardosin a);
PDBTitle: native cardosin a from cynara cardunculus l.
|
| 79 | d1dx5i2 |
|
not modelled |
8.6 |
29 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:EGF/Laminin
Family:EGF-type module |
| 80 | d1g5ga1 |
|
not modelled |
8.5 |
34 |
Fold:Head and neck region of the ectodomain of NDV fusion glycoprotein
Superfamily:Head and neck region of the ectodomain of NDV fusion glycoprotein
Family:Head and neck region of the ectodomain of NDV fusion glycoprotein |
| 81 | c3g6nA_ |
|
not modelled |
8.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: crystal structure of an efpdf complex with met-ala-ser
|
| 82 | c3pilA_ |
|
not modelled |
8.3 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase;
PDBTitle: crystal structure of mxr1 from saccharomyces cerevisiae in reduced2 form
|
| 83 | c1ciiA_ |
|
not modelled |
8.2 |
7 |
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
|
| 84 | c3bzjA_ |
|
not modelled |
8.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:uv endonuclease;
PDBTitle: uvde k229l
|
| 85 | c2kppA_ |
|
not modelled |
8.2 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin0431 protein;
PDBTitle: solution nmr structure of lin0431 protein from listeria innocua.2 northeast structural genomics consortium target lkr112
|
| 86 | c3ry6A_ |
|
not modelled |
8.2 |
21 |
PDB header:immune system
Chain: A: PDB Molecule:ig gamma-1 chain c region;
PDBTitle: complex of fcgammariia (cd32) and the fc of human igg1
|
| 87 | c3ry6B_ |
|
not modelled |
8.2 |
21 |
PDB header:immune system
Chain: B: PDB Molecule:ig gamma-1 chain c region;
PDBTitle: complex of fcgammariia (cd32) and the fc of human igg1
|
| 88 | d1e88a3 |
|
not modelled |
8.1 |
21 |
Fold:FnI-like domain
Superfamily:FnI-like domain
Family:Fibronectin type I module |
| 89 | c2k5lA_ |
|
not modelled |
8.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:feoa;
PDBTitle: solution nmr structure of protein feoa from clostridium2 thermocellum, northeast structural genomics consortium3 target cmr17
|
| 90 | d1kshb_ |
|
not modelled |
8.0 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:RhoGDI-like |
| 91 | d1xdna_ |
|
not modelled |
8.0 |
22 |
Fold:ATP-grasp
Superfamily:DNA ligase/mRNA capping enzyme, catalytic domain
Family:RNA ligase |
| 92 | d1jv2a2 |
|
not modelled |
7.9 |
22 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Integrin domains
Family:Integrin domains |
| 93 | c1no7A_ |
|
not modelled |
7.9 |
15 |
PDB header:viral protein
Chain: A: PDB Molecule:major capsid protein;
PDBTitle: structure of the large protease resistant upper domain of2 vp5, the major capsid protein of herpes simplex virus-1
|
| 94 | d1no7a_ |
|
not modelled |
7.9 |
15 |
Fold:Major capsid protein VP5
Superfamily:Major capsid protein VP5
Family:Major capsid protein VP5 |
| 95 | c1oqzB_ |
|
not modelled |
7.9 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:glutaryl acylase;
PDBTitle: crystal structures of glutaryl 7-aminocephalosporanic acid acylase:2 insight into autoproteolytic activation
|
| 96 | d1hica_ |
|
not modelled |
7.9 |
69 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Leech antihemostatic proteins
Family:Hirudin-like |
| 97 | d1shwb_ |
|
not modelled |
7.9 |
29 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Ephrin receptor ligand binding domain |
| 98 | c1rh5C_ |
|
not modelled |
7.7 |
39 |
PDB header:protein transport
Chain: C: PDB Molecule:secbeta;
PDBTitle: the structure of a protein conducting channel
|
| 99 | d1rh5c_ |
|
not modelled |
7.7 |
39 |
Fold:Single transmembrane helix
Superfamily:Sec-beta subunit
Family:Sec-beta subunit |