1 c2x3yA_
100.0
40
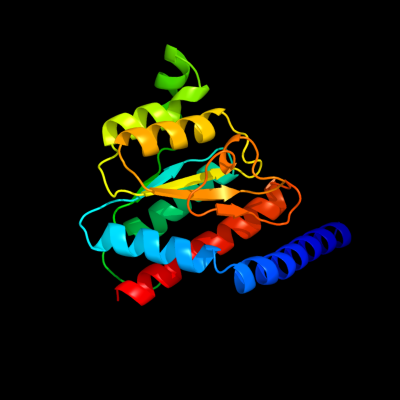
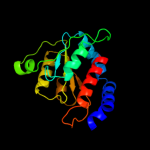
PDB header: isomeraseChain: A: PDB Molecule: phosphoheptose isomerase;PDBTitle: crystal structure of gmha from burkholderia pseudomallei
2 d1tk9a_
100.0
44
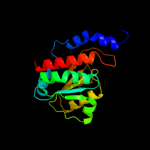
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain3 c2yvaB_
100.0
40
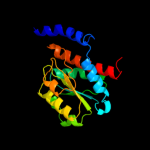
PDB header: dna binding proteinChain: B: PDB Molecule: dnaa initiator-associating protein diaa;PDBTitle: crystal structure of escherichia coli diaa
4 d1x92a_
100.0
47
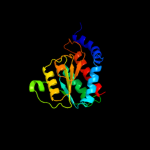
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain5 d1x94a_
100.0
80
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain6 c3trjC_
100.0
39
PDB header: isomeraseChain: C: PDB Molecule: phosphoheptose isomerase;PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
7 c1nriA_
99.9
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein hi0754;PDBTitle: crystal structure of putative phosphosugar isomerase hi0754 from2 haemophilus influenzae
8 d1nria_
99.9
19
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain9 c3cvjB_
99.9
21
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
10 c3fxaA_
99.9
22
PDB header: sugar binding proteinChain: A: PDB Molecule: sis domain protein;PDBTitle: crystal structure of a putative sugar-phosphate isomerase2 (lmof2365_0531) from listeria monocytogenes str. 4b f2365 at 1.60 a3 resolution
11 c3etnD_
99.8
21
PDB header: isomeraseChain: D: PDB Molecule: putative phosphosugar isomerase involved in capsulePDBTitle: crystal structure of putative phosphosugar isomerase involved in2 capsule formation (yp_209877.1) from bacteroides fragilis nctc 93433 at 1.70 a resolution
12 c3shoA_
99.8
22
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, rpir family;PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
13 c2xhzC_
99.8
22
PDB header: isomeraseChain: C: PDB Molecule: arabinose 5-phosphate isomerase;PDBTitle: probing the active site of the sugar isomerase domain from e. coli2 arabinose-5-phosphate isomerase via x-ray crystallography
14 d1vima_
99.8
23
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain15 d1m3sa_
99.8
21
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain16 c3hbaA_
99.8
17
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerase;PDBTitle: crystal structure of a putative phosphosugar isomerase (sden_2705)2 from shewanella denitrificans os217 at 2.00 a resolution
17 c2zj3A_
99.8
14
PDB header: transferaseChain: A: PDB Molecule: glucosamine--fructose-6-phosphatePDBTitle: isomerase domain of human glucose:fructose-6-phosphate2 amidotransferase
18 c3knzA_
99.8
18
PDB header: sugar binding proteinChain: A: PDB Molecule: putative sugar binding protein;PDBTitle: crystal structure of putative sugar binding protein (np_459565.1) from2 salmonella typhimurium lt2 at 2.50 a resolution
19 c3fj1A_
99.8
17
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerase;PDBTitle: crystal structure of putative phosphosugar isomerase (yp_167080.1)2 from silicibacter pomeroyi dss-3 at 1.75 a resolution
20 c2puwA_
99.8
13
PDB header: transferaseChain: A: PDB Molecule: isomerase domain of glutamine-fructose-6-phosphatePDBTitle: the crystal structure of isomerase domain of glucosamine-6-phosphate2 synthase from candida albicans
21 d1jeoa_
not modelled
99.8
19
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain22 c2a3nA_
not modelled
99.7
13
PDB header: sugar binding proteinChain: A: PDB Molecule: putative glucosamine-fructose-6-phosphate aminotransferase;PDBTitle: crystal structure of a putative glucosamine-fructose-6-phosphate2 aminotransferase (stm4540.s) from salmonella typhimurium lt2 at 1.353 a resolution
23 c3g68A_
not modelled
99.7
16
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerase;PDBTitle: crystal structure of a putative phosphosugar isomerase (cd3275) from2 clostridium difficile 630 at 1.80 a resolution
24 d1moqa_
not modelled
99.7
14
Fold: SIS domainSuperfamily: SIS domainFamily: double-SIS domain25 c2amlB_
not modelled
99.7
24
PDB header: transferaseChain: B: PDB Molecule: sis domain protein;PDBTitle: crystal structure of lmo0035 protein (46906266) from listeria2 monocytogenes 4b f2365 at 1.50 a resolution
26 c3euaD_
not modelled
99.7
21
PDB header: isomeraseChain: D: PDB Molecule: putative fructose-aminoacid-6-phosphate deglycase;PDBTitle: crystal structure of a putative phosphosugar isomerase (bsu32610) from2 bacillus subtilis at 1.90 a resolution
27 d1j5xa_
not modelled
99.7
18
Fold: SIS domainSuperfamily: SIS domainFamily: double-SIS domain28 c3tbfA_
not modelled
99.7
14
PDB header: transferaseChain: A: PDB Molecule: glucosamine--fructose-6-phosphate aminotransferasePDBTitle: c-terminal domain of glucosamine-fructose-6-phosphate aminotransferase2 from francisella tularensis.
29 c1jxaA_
not modelled
99.6
14
PDB header: transferaseChain: A: PDB Molecule: glucosamine 6-phosphate synthase;PDBTitle: glucosamine 6-phosphate synthase with glucose 6-phosphate
30 c3fkjA_
not modelled
99.6
18
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerases;PDBTitle: crystal structure of a putative phosphosugar isomerase (stm_0572) from2 salmonella typhimurium lt2 at 2.12 a resolution
31 c3i0zB_
not modelled
99.5
20
PDB header: isomeraseChain: B: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of putative putative tagatose-6-phosphate2 ketose/aldose isomerase (np_344614.1) from streptococcus pneumoniae3 tigr4 at 1.70 a resolution
32 d1x9ia_
not modelled
99.5
20
Fold: SIS domainSuperfamily: SIS domainFamily: double-SIS domain33 c3odpA_
not modelled
99.5
17
PDB header: isomeraseChain: A: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of a putative tagatose-6-phosphate ketose/aldose2 isomerase (nt01cx_0292) from clostridium novyi nt at 2.35 a3 resolution
34 c2decA_
not modelled
99.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 325aa long hypothetical protein;PDBTitle: crystal structure of the ph0510 protein from pyrococcus horikoshii ot3
35 c3c3jA_
not modelled
99.5
17
PDB header: isomeraseChain: A: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
36 c3jx9B_
not modelled
98.2
11
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of putative phosphoheptose isomerase2 (yp_001815198.1) from exiguobacterium sp. 255-15 at 1.95 a resolution
37 c3ff1B_
not modelled
98.2
21
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: structure of glucose 6-phosphate isomerase from staphylococcus aureus
38 c2q8nB_
not modelled
98.1
18
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of glucose-6-phosphate isomerase (ec2 5.3.1.9) (tm1385) from thermotoga maritima at 1.82 a3 resolution
39 d1c7qa_
not modelled
98.1
17
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI40 c1zzgB_
not modelled
97.7
15
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of hypothetical protein tt0462 from thermus2 thermophilus hb8
41 c3iz6A_
not modelled
97.5
15
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein sa (s2p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
42 d2gy9b1
not modelled
97.2
17
Fold: Flavodoxin-likeSuperfamily: Ribosomal protein S2Family: Ribosomal protein S243 d2uubb1
not modelled
97.2
20
Fold: Flavodoxin-likeSuperfamily: Ribosomal protein S2Family: Ribosomal protein S244 c2zkqb_
not modelled
97.1
14
PDB header: ribosomal protein/rnaChain: B: PDB Molecule: rna expansion segment es3;PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
45 c3bbnB_
not modelled
97.1
13
PDB header: ribosomeChain: B: PDB Molecule: ribosomal protein s2;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
46 d1gzda_
not modelled
96.9
20
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI47 c3hjbA_
not modelled
96.8
21
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: 1.5 angstrom crystal structure of glucose-6-phosphate isomerase from2 vibrio cholerae.
48 d1iata_
not modelled
96.8
18
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI49 c1s1hB_
not modelled
96.7
18
PDB header: ribosomeChain: B: PDB Molecule: 40s ribosomal protein s0-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
50 c3izbA_
not modelled
96.7
18
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein rps0 (s2p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
51 c2xznB_
not modelled
96.7
18
PDB header: ribosomeChain: B: PDB Molecule: rps0e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
52 d2jioa2
not modelled
96.6
16
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-353 c3ujhB_
not modelled
96.6
19
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of substrate-bound glucose-6-phosphate isomerase2 from toxoplasma gondii
54 d1h0ha2
not modelled
96.6
13
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-355 c2wu8A_
not modelled
96.6
17
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: structural studies of phosphoglucose isomerase from2 mycobacterium tuberculosis h37rv
56 c2v45A_
not modelled
96.5
15
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic nitrate reductase;PDBTitle: a new catalytic mechanism of periplasmic nitrate reductase2 from desulfovibrio desulfuricans atcc 27774 from3 crystallographic and epr data and based on detailed4 analysis of the sixth ligand
57 c3ljkA_
not modelled
96.5
19
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: glucose-6-phosphate isomerase from francisella tularensis.
58 d1u0fa_
not modelled
96.5
20
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI59 c2e7zA_
not modelled
96.5
17
PDB header: lyaseChain: A: PDB Molecule: acetylene hydratase ahy;PDBTitle: acetylene hydratase from pelobacter acetylenicus
60 c3bchA_
not modelled
96.5
14
PDB header: cell adhesion, ribosomal proteinChain: A: PDB Molecule: 40s ribosomal protein sa;PDBTitle: crystal structure of the human laminin receptor precursor
61 c1y5iA_
not modelled
96.4
10
PDB header: oxidoreductaseChain: A: PDB Molecule: respiratory nitrate reductase 1 alpha chain;PDBTitle: the crystal structure of the narghi mutant nari-k86a
62 d1hm5a_
not modelled
96.3
18
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI63 d2iv2x2
not modelled
96.2
16
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-364 d1vi6a_
not modelled
96.2
19
Fold: Flavodoxin-likeSuperfamily: Ribosomal protein S2Family: Ribosomal protein S265 c1h0hA_
not modelled
96.2
11
PDB header: dehydrogenaseChain: A: PDB Molecule: formate dehydrogenase (large subunit);PDBTitle: tungsten containing formate dehydrogenase from2 desulfovibrio gigas
66 c3nbuC_
not modelled
96.1
22
PDB header: isomeraseChain: C: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of pgi glucosephosphate isomerase
67 c2nyaF_
not modelled
96.1
13
PDB header: oxidoreductaseChain: F: PDB Molecule: periplasmic nitrate reductase;PDBTitle: crystal structure of the periplasmic nitrate reductase2 (nap) from escherichia coli
68 c2ivfA_
not modelled
95.9
16
PDB header: oxidoreductaseChain: A: PDB Molecule: ethylbenzene dehydrogenase alpha-subunit;PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
69 d1ogya2
not modelled
95.9
14
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-370 c2vpyE_
not modelled
95.9
14
PDB header: oxidoreductaseChain: E: PDB Molecule: thiosulfate reductase;PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
71 c1ogyA_
not modelled
95.9
14
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic nitrate reductase;PDBTitle: crystal structure of the heterodimeric nitrate reductase2 from rhodobacter sphaeroides
72 d1y5ia2
not modelled
95.9
11
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-373 d1kqfa2
not modelled
95.8
13
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-374 c2o2cB_
not modelled
95.8
20
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase, glycosomal;PDBTitle: crystal structure of phosphoglucose isomerase from t. brucei2 containing glucose-6-phosphate in the active site
75 c1t10A_
not modelled
95.8
16
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: phosphoglucose isomerase from leishmania mexicana in complex with2 substrate d-fructose-6-phosphate
76 d1q50a_
not modelled
95.7
16
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI77 c3pr3B_
not modelled
95.6
18
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of plasmodium falciparum glucose-6-phosphate2 isomerase (pf14_0341) in complex with fructose-6-phosphate
78 c1h5nC_
not modelled
95.5
11
PDB header: oxidoreductaseChain: C: PDB Molecule: dmso reductase;PDBTitle: dmso reductase modified by the presence of dms and air
79 d1tmoa2
not modelled
95.2
10
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-380 c1tmoA_
not modelled
95.2
9
PDB header: oxidoreductaseChain: A: PDB Molecule: trimethylamine n-oxide reductase;PDBTitle: trimethylamine n-oxide reductase from shewanella massilia
81 d1dmra2
not modelled
95.1
12
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-382 c2iv2X_
not modelled
95.0
13
PDB header: oxidoreductaseChain: X: PDB Molecule: formate dehydrogenase h;PDBTitle: reinterpretation of reduced form of formate dehydrogenase h2 from e. coli
83 c1kqgA_
not modelled
94.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: formate dehydrogenase, nitrate-inducible, major subunit;PDBTitle: formate dehydrogenase n from e. coli
84 d1vlfm2
not modelled
93.9
9
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-385 c1vlfQ_
not modelled
93.8
9
PDB header: oxidoreductaseChain: Q: PDB Molecule: pyrogallol hydroxytransferase large subunit;PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici complexed3 with inhibitor 1,2,4,5-tetrahydroxy-benzene
86 d1p3da1
not modelled
91.7
16
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain87 d1m2ka_
not modelled
91.0
14
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators88 d2b4ya1
not modelled
90.7
10
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators89 c3jwpA_
not modelled
90.3
14
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein sir2 homologue;PDBTitle: crystal structure of plasmodium falciparum sir2a (pf13_0152) in2 complex with amp
90 d1yc5a1
not modelled
90.2
11
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators91 c1ir6A_
not modelled
89.9
16
PDB header: hydrolaseChain: A: PDB Molecule: exonuclease recj;PDBTitle: crystal structure of exonuclease recj bound to manganese
92 d1ir6a_
not modelled
89.9
16
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Exonuclease RecJ93 d1j6ua1
not modelled
89.7
15
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain94 c3k35D_
not modelled
89.5
16
PDB header: hydrolaseChain: D: PDB Molecule: nad-dependent deacetylase sirtuin-6;PDBTitle: crystal structure of human sirt6
95 d1ma3a_
not modelled
88.5
12
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators96 c3pkiF_
not modelled
88.5
16
PDB header: hydrolaseChain: F: PDB Molecule: nad-dependent deacetylase sirtuin-6;PDBTitle: human sirt6 crystal structure in complex with adp ribose
97 c3uagA_
not modelled
88.0
15
PDB header: ligaseChain: A: PDB Molecule: protein (udp-n-acetylmuramoyl-l-alanine:d-PDBTitle: udp-n-acetylmuramoyl-l-alanine:d-glutamate ligase
98 c3ndjA_
not modelled
87.9
9
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: x-ray structure of a c-3'-methyltransferase in complex with s-2 adenosyl-l-homocysteine and sugar product
99 d1g8ka2
not modelled
87.2
11
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3100 c3q2oB_
not modelled
86.1
25
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
101 c1g8jC_
not modelled
86.0
12
PDB header: oxidoreductaseChain: C: PDB Molecule: arsenite oxidase;PDBTitle: crystal structure analysis of arsenite oxidase from2 alcaligenes faecalis
102 d1eu1a2
not modelled
85.8
8
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3103 d1kjqa2
not modelled
85.4
10
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like104 d2ax3a2
not modelled
84.7
15
Fold: YjeF N-terminal domain-likeSuperfamily: YjeF N-terminal domain-likeFamily: YjeF N-terminal domain-like105 c1j6uA_
not modelled
84.5
15
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-alanine ligase murc;PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase2 murc (tm0231) from thermotoga maritima at 2.3 a resolution
106 c3uvzB_
not modelled
84.3
19
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase, atpase2 subunit from burkholderia ambifaria
107 c3ouzA_
not modelled
84.3
18
PDB header: ligaseChain: A: PDB Molecule: biotin carboxylase;PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
108 c1yfzA_
not modelled
84.1
10
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis
109 d1yfza1
not modelled
84.1
10
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)110 d2jfga1
not modelled
84.0
13
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain111 d1a9xa4
not modelled
83.4
21
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like112 c2dwcB_
not modelled
83.4
13
PDB header: transferaseChain: B: PDB Molecule: 433aa long hypothetical phosphoribosylglycinamide formylPDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
113 d1rq2a1
not modelled
82.4
21
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain114 c1y7pB_
not modelled
82.3
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein af1403;PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
115 c3orqA_
not modelled
81.6
17
PDB header: ligase,biosynthetic proteinChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide synthetase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
116 c2z04A_
not modelled
81.5
24
PDB header: lyaseChain: A: PDB Molecule: phosphoribosylaminoimidazole carboxylase atpasePDBTitle: crystal structure of phosphoribosylaminoimidazole2 carboxylase atpase subunit from aquifex aeolicus
117 c1eu1A_
not modelled
81.3
11
PDB header: oxidoreductaseChain: A: PDB Molecule: dimethyl sulfoxide reductase;PDBTitle: the crystal structure of rhodobacter sphaeroides dimethylsulfoxide2 reductase reveals two distinct molybdenum coordination environments.
118 c1kjjA_
not modelled
80.6
9
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase 2;PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
119 c2q1yB_
not modelled
79.2
20
PDB header: cell cycle, signaling proteinChain: B: PDB Molecule: cell division protein ftsz;PDBTitle: crystal structure of cell division protein ftsz from mycobacterium2 tuberculosis in complex with gtp-gamma-s
120 c2vpqA_
not modelled
79.2
21
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase;PDBTitle: crystal structure of biotin carboxylase from s. aureus2 complexed with amppnp