1 c2vv5D_
100.0
20
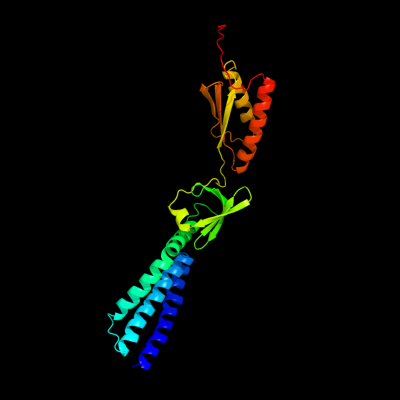
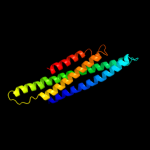
PDB header: membrane proteinChain: D: PDB Molecule: small-conductance mechanosensitive channel;PDBTitle: the open structure of mscs
2 d2vv5a2
99.6
20
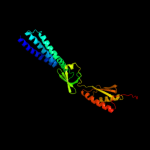
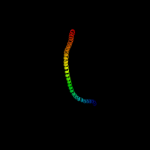
Fold: Ferredoxin-likeSuperfamily: Mechanosensitive channel protein MscS (YggB), C-terminal domainFamily: Mechanosensitive channel protein MscS (YggB), C-terminal domain3 d2vv5a1
99.2
29
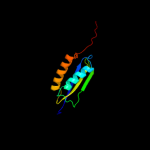
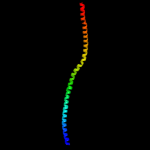
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Mechanosensitive channel protein MscS (YggB), middle domain4 d2vv5a3
98.7
12
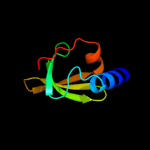
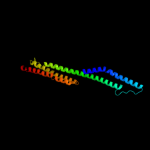
Fold: Mechanosensitive channel protein MscS (YggB), transmembrane regionSuperfamily: Mechanosensitive channel protein MscS (YggB), transmembrane regionFamily: Mechanosensitive channel protein MscS (YggB), transmembrane region5 c1ciiA_
98.7
12
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
6 c1c1gA_
98.5
12
PDB header: contractile proteinChain: A: PDB Molecule: tropomyosin;PDBTitle: crystal structure of tropomyosin at 7 angstroms resolution2 in the spermine-induced crystal form
7 c1yvlB_
98.1
9
PDB header: signaling proteinChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: structure of unphosphorylated stat1
8 c1sjjB_
97.9
10
PDB header: contractile proteinChain: B: PDB Molecule: actinin;PDBTitle: cryo-em structure of chicken gizzard smooth muscle alpha-2 actinin
9 c2oevA_
97.7
9
PDB header: protein transportChain: A: PDB Molecule: programmed cell death 6-interacting protein;PDBTitle: crystal structure of alix/aip1
10 c1bf5A_
97.7
9
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: tyrosine phosphorylated stat-1/dna complex
11 c3ojaB_
97.5
12
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
12 c2oexB_
97.3
8
PDB header: protein transportChain: B: PDB Molecule: programmed cell death 6-interacting protein;PDBTitle: structure of alix/aip1 v domain
13 c1bg1A_
97.2
10
PDB header: transcription/dnaChain: A: PDB Molecule: protein (transcription factor stat3b);PDBTitle: transcription factor stat3b/dna complex
14 c2efrB_
97.0
7
PDB header: contractile proteinChain: B: PDB Molecule: general control protein gcn4 and tropomyosin 1 alpha chain;PDBTitle: crystal structure of the c-terminal tropomyosin fragment with n- and2 c-terminal extensions of the leucine zipper at 1.8 angstroms3 resolution
15 c3na7A_
96.9
10
PDB header: gene regulation, chaperoneChain: A: PDB Molecule: hp0958;PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
16 c3cwgA_
96.9
10
PDB header: transcriptionChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: unphosphorylated mouse stat3 core fragment
17 c3dtpA_
96.5
12
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
18 c1f5nA_
96.4
8
PDB header: signaling proteinChain: A: PDB Molecule: interferon-induced guanylate-binding protein 1;PDBTitle: human guanylate binding protein-1 in complex with the gtp2 analogue, gmppnp.
19 c1ei3C_
96.4
8
PDB header: PDB COMPND: 20 c1g8xB_
96.4
9
PDB header: structural proteinChain: B: PDB Molecule: myosin ii heavy chain fused to alpha-actinin 3;PDBTitle: structure of a genetically engineered molecular motor
21 c3ghgK_
not modelled
96.1
9
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
22 c3hizB_
not modelled
95.9
10
PDB header: transferase/oncoproteinChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunitPDBTitle: crystal structure of p110alpha h1047r mutant in complex with2 nish2 of p85alpha
23 c2d3eD_
not modelled
95.6
9
PDB header: contractile proteinChain: D: PDB Molecule: general control protein gcn4 and tropomyosin 1PDBTitle: crystal structure of the c-terminal fragment of rabbit2 skeletal alpha-tropomyosin
24 c2v71A_
not modelled
95.6
9
PDB header: nuclear proteinChain: A: PDB Molecule: nuclear distribution protein nude-like 1;PDBTitle: coiled-coil region of nudel
25 c2ch7A_
not modelled
95.4
10
PDB header: chemotaxisChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of the cytoplasmic domain of a bacterial2 chemoreceptor from thermotoga maritima
26 c1jchC_
not modelled
95.3
4
PDB header: ribosome inhibitor, hydrolaseChain: C: PDB Molecule: colicin e3;PDBTitle: crystal structure of colicin e3 in complex with its immunity protein
27 c1ei3E_
not modelled
95.0
11
PDB header: PDB COMPND: 28 c3o0zD_
not modelled
94.9
14
PDB header: transferaseChain: D: PDB Molecule: rho-associated protein kinase 1;PDBTitle: crystal structure of a coiled-coil domain from human rock i
29 c2fxmB_
not modelled
94.8
13
PDB header: contractile proteinChain: B: PDB Molecule: myosin heavy chain, cardiac muscle beta isoform;PDBTitle: structure of the human beta-myosin s2 fragment
30 c3ojaA_
not modelled
94.7
11
PDB header: protein bindingChain: A: PDB Molecule: leucine-rich immune molecule 1;PDBTitle: crystal structure of lrim1/apl1c complex
31 c4a55B_
not modelled
94.7
8
PDB header: transferaseChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit alpha;PDBTitle: crystal structure of p110alpha in complex with ish2 of p85alpha and2 the inhibitor pik-108
32 c1y4cA_
not modelled
94.7
12
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
33 c1deqF_
not modelled
94.5
5
PDB header: PDB COMPND: 34 c2dfsA_
not modelled
94.1
10
PDB header: contractile protein/transport proteinChain: A: PDB Molecule: myosin-5a;PDBTitle: 3-d structure of myosin-v inhibited state
35 c2v1yB_
not modelled
94.0
7
PDB header: transferaseChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit alpha;PDBTitle: structure of a phosphoinositide 3-kinase alpha adaptor-2 binding domain (abd) in a complex with the ish2 domain3 from p85 alpha
36 c2y3aB_
not modelled
93.9
13
PDB header: transferaseChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit beta;PDBTitle: crystal structure of p110beta in complex with icsh2 of p85beta and2 the drug gdc-0941
37 c1deqO_
not modelled
93.8
9
PDB header: PDB COMPND: 38 c2b9cA_
not modelled
93.6
12
PDB header: contractile proteinChain: A: PDB Molecule: striated-muscle alpha tropomyosin;PDBTitle: structure of tropomyosin's mid-region: bending and binding2 sites for actin
39 c2j69D_
not modelled
92.1
10
PDB header: hydrolaseChain: D: PDB Molecule: bacterial dynamin-like protein;PDBTitle: bacterial dynamin-like protein bdlp
40 c3u59C_
not modelled
91.2
15
PDB header: contractile proteinChain: C: PDB Molecule: tropomyosin beta chain;PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
41 c1hciB_
not modelled
91.1
11
PDB header: triple-helix coiled coilChain: B: PDB Molecule: alpha-actinin 2;PDBTitle: crystal structure of the rod domain of alpha-actinin
42 c3ipkA_
not modelled
89.6
12
PDB header: cell adhesionChain: A: PDB Molecule: agi/ii;PDBTitle: crystal structure of a3vp1 of agi/ii of streptococcus mutans
43 c1deqD_
not modelled
87.8
10
PDB header: PDB COMPND: 44 d2ap3a1
not modelled
87.4
11
Fold: Four-helical up-and-down bundleSuperfamily: MW0975(SA0943)-likeFamily: MW0975(SA0943)-like45 c3hnwB_
not modelled
84.8
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
46 c2zkrt_
not modelled
84.8
24
PDB header: ribosomal protein/rnaChain: T: PDB Molecule: rna expansion segment es39 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
47 c3ol1A_
not modelled
84.8
12
PDB header: structural proteinChain: A: PDB Molecule: vimentin;PDBTitle: crystal structure of vimentin (fragment 144-251) from homo sapiens,2 northeast structural genomics consortium target hr4796b
48 c2v66C_
not modelled
84.5
11
PDB header: structural proteinChain: C: PDB Molecule: nuclear distribution protein nude-like 1;PDBTitle: crystal structure of the coiled-coil domain of ndel1 (a.a.2 58 to 169)c
49 c3r6nA_
not modelled
84.1
5
PDB header: cell adhesionChain: A: PDB Molecule: desmoplakin;PDBTitle: crystal structure of a rigid four spectrin repeat fragment of the2 human desmoplakin plakin domain
50 c2gl2B_
not modelled
83.5
10
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
51 c3g67A_
not modelled
83.3
7
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of a soluble chemoreceptor from thermotoga2 maritima
52 c3l9oA_
not modelled
80.6
13
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: crystal structure of mtr4, a co-factor of the nuclear exosome
53 d2hqha1
not modelled
80.5
33
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain54 d2cqaa1
not modelled
79.8
32
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TIP49 domain55 c2rd0B_
not modelled
77.6
10
PDB header: transferase/oncoproteinChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit alpha;PDBTitle: structure of a human p110alpha/p85alpha complex
56 c3edvB_
not modelled
76.3
10
PDB header: structural proteinChain: B: PDB Molecule: spectrin beta chain, brain 1;PDBTitle: crystal structure of repeats 14-16 of beta2-spectrin
57 c1l8dB_
not modelled
74.4
7
PDB header: replicationChain: B: PDB Molecule: dna double-strand break repair rad50 atpase;PDBTitle: rad50 coiled-coil zn hook
58 c2zv4O_
not modelled
74.3
13
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
59 d2cp6a1
not modelled
74.2
30
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain60 d1nz9a_
not modelled
74.0
35
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain61 c3kbtA_
not modelled
73.9
9
PDB header: structural proteinChain: A: PDB Molecule: spectrin beta chain, erythrocyte;PDBTitle: crystal structure of the ankyrin binding domain of human erythroid2 beta spectrin (repeats 13-15) in complex with the spectrin binding3 domain of human erythroid ankyrin (zu5-ank)
62 d1nppa2
not modelled
72.0
22
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain63 c2wpqA_
not modelled
71.6
9
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
64 c2kvqG_
not modelled
70.9
18
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
65 c2jvvA_
not modelled
70.9
18
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
66 c4a1cS_
not modelled
70.5
40
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
67 d1vqot1
not modelled
68.6
15
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e68 c3iz5Y_
not modelled
67.0
35
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
69 c3lssA_
not modelled
66.5
13
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: trypanosoma brucei seryl-trna synthetase in complex with atp
70 c2e6zA_
not modelled
66.0
20
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
71 d1whka_
not modelled
65.9
13
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain72 d2plsa1
not modelled
65.2
17
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like73 c2vfyA_
not modelled
65.0
16
PDB header: hydrolaseChain: A: PDB Molecule: akap18 delta;PDBTitle: akap18 delta central domain
74 d1whma_
not modelled
63.3
23
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain75 d2coya1
not modelled
63.2
27
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain76 c2i1jA_
not modelled
62.8
11
PDB header: cell adhesion, membrane proteinChain: A: PDB Molecule: moesin;PDBTitle: moesin from spodoptera frugiperda at 2.1 angstroms resolution
77 d2cp5a1
not modelled
62.5
20
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain78 d1t9ha1
not modelled
61.5
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like79 d2coza1
not modelled
60.8
27
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain80 c2e7sM_
not modelled
60.8
7
PDB header: endocytosis/exocytosisChain: M: PDB Molecule: rab guanine nucleotide exchange factor sec2;PDBTitle: crystal structure of the yeast sec2p gef domain
81 c1i84V_
not modelled
60.6
18
PDB header: contractile proteinChain: V: PDB Molecule: smooth muscle myosin heavy chain;PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.
82 c1l2fA_
not modelled
60.6
19
PDB header: transcriptionChain: A: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima: a2 structure-based role of the n-terminal domain
83 d2cp2a1
not modelled
60.5
20
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain84 c2qihA_
not modelled
60.5
15
PDB header: cell adhesionChain: A: PDB Molecule: protein uspa1;PDBTitle: crystal structure of 527-665 fragment of uspa1 protein from2 moraxella catarrhalis
85 c3cnrA_
not modelled
59.6
23
PDB header: unknown functionChain: A: PDB Molecule: type iv fimbriae assembly protein;PDBTitle: crystal structure of pilz (xac1133) from xanthomonas2 axonopodis pv citri
86 d1yeza1
not modelled
59.5
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain87 c2jeeA_
not modelled
59.5
9
PDB header: cell cycleChain: A: PDB Molecule: yiiu;PDBTitle: xray structure of e. coli yiiu
88 d1whja_
not modelled
59.4
24
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain89 d2e3ha1
not modelled
59.4
30
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain90 c2k52A_
not modelled
58.8
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mj1198;PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
91 d2do3a1
not modelled
57.1
28
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like92 d2p13a1
not modelled
57.0
17
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like93 c2no2A_
not modelled
56.9
12
PDB header: cell adhesionChain: A: PDB Molecule: huntingtin-interacting protein 1;PDBTitle: crystal structure of the dllrkn-containing coiled-coil2 domain of huntingtin-interacting protein 1
94 c2e4hA_
not modelled
56.6
27
PDB header: structural proteinChain: A: PDB Molecule: restin;PDBTitle: solution structure of cytoskeletal protein in complex with2 tubulin tail
95 d2e3ia1
not modelled
56.3
20
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain96 c2rcnA_
not modelled
55.3
21
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of the ribosomal interacting gtpase yjeq from the2 enterobacterial species salmonella typhimurium.
97 c1e0tD_
not modelled
55.2
18
PDB header: phosphotransferaseChain: D: PDB Molecule: pyruvate kinase;PDBTitle: r292d mutant of e. coli pyruvate kinase
98 c2zdiA_
not modelled
55.1
12
PDB header: chaperoneChain: A: PDB Molecule: prefoldin subunit beta;PDBTitle: crystal structure of prefoldin from pyrococcus horikoshii2 ot3
99 d2cp3a1
not modelled
54.5
35
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain100 d1h6za2
not modelled
54.3
38
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain101 d1vqoq1
not modelled
54.1
27
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e102 c2p03A_
not modelled
54.1
10
PDB header: cell adhesionChain: A: PDB Molecule: alpha-2-macroglobulin receptor-associatedPDBTitle: the structure of receptor-associated protein(rap)
103 c2z0wA_
not modelled
53.7
20
PDB header: protein bindingChain: A: PDB Molecule: cap-gly domain-containing linker protein 4;PDBTitle: crystal structure of the 2nd cap-gly domain in human restin-2 like protein 2 reveals a swapped-dimer
104 d2cowa1
not modelled
53.7
32
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain105 c1q46A_
not modelled
53.3
18
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
106 c2v4hA_
not modelled
52.9
12
PDB header: transcriptionChain: A: PDB Molecule: nf-kappa-b essential modulator;PDBTitle: nemo cc2-lz domain - 1d5 darpin complex
107 c3b7kA_
not modelled
52.7
21
PDB header: hydrolaseChain: A: PDB Molecule: acyl-coenzyme a thioesterase 12;PDBTitle: human acyl-coenzyme a thioesterase 12
108 c2wctC_
not modelled
52.7
37
PDB header: rna-binding proteinChain: C: PDB Molecule: non-structural protein 3;PDBTitle: human sars coronavirus unique domain (triclinic form)
109 d2essa1
not modelled
52.1
15
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: Acyl-ACP thioesterase-like110 c1hh2P_
not modelled
52.0
20
PDB header: transcription regulationChain: P: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima
111 d1yvca1
not modelled
51.9
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain112 d1ppje2
not modelled
51.5
22
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor113 c3iz5U_
not modelled
51.4
24
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
114 d2o1ra1
not modelled
51.3
21
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like115 d1whha_
not modelled
51.3
24
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain116 c4a1aP_
not modelled
51.2
28
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l21;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
117 c3a7pB_
not modelled
50.9
8
PDB header: protein transportChain: B: PDB Molecule: autophagy protein 16;PDBTitle: the crystal structure of saccharomyces cerevisiae atg16
118 d2f9ha1
not modelled
50.8
26
Fold: PTSIIA/GutA-likeSuperfamily: PTSIIA/GutA-likeFamily: PTSIIA/GutA-like119 c3dedB_
not modelled
50.4
8
PDB header: membrane proteinChain: B: PDB Molecule: probable hemolysin;PDBTitle: c-terminal domain of probable hemolysin from chromobacterium violaceum
120 d1y7ua1
not modelled
50.4
13
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like