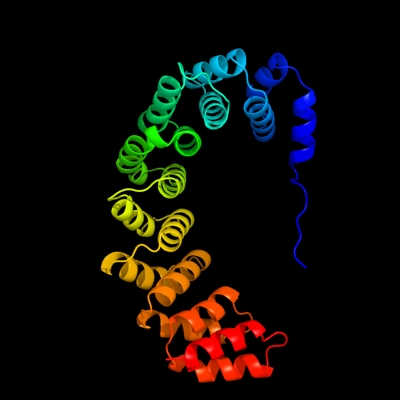
| 1 | d1oyza_
|
|
 |
100.0 |
99 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:PBS lyase HEAT-like repeat |
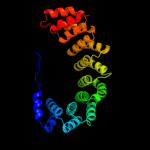
| 2 | c3ltjA_
|
|
 |
99.9 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:alpharep-4;
PDBTitle: structure of a new family of artificial alpha helicoidal repeat2 proteins (alpha-rep) based on thermostable heat-like repeats
|
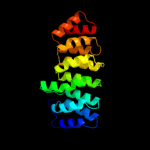
| 3 | c2db0B_
|
|
 |
99.7 |
17 |
PDB header:protein binding
Chain: B: PDB Molecule:253aa long hypothetical protein;
PDBTitle: crystal structure of ph0542
|
| 4 | d1gw5b_
|
|
 |
99.7 |
13 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Clathrin adaptor core protein |
| 5 | c3c5wA_
|
|
 |
99.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:pp2a a subunit;
PDBTitle: complex between pp2a-specific methylesterase pme-1 and pp2a core2 enzyme
|
| 6 | c3l6yA_
|
|
 |
99.5 |
11 |
PDB header:cell adhesion
Chain: A: PDB Molecule:catenin delta-1;
PDBTitle: crystal structure of p120 catenin in complex with e-cadherin
|
| 7 | d1te4a_
|
|
 |
99.5 |
17 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:PBS lyase HEAT-like repeat |
| 8 | c3b2aA_
|
|
 |
99.5 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the archaeal heat-like repeats protein ton_19372 from thermococcus onnurineus na1
|
| 9 | c3l9tA_
|
|
 |
99.5 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein smu.31;
PDBTitle: the crystal structure of smu.31 from streptococcus mutans ua159
|
| 10 | c1w63A_
|
|
 |
99.4 |
13 |
PDB header:endocytosis
Chain: A: PDB Molecule:adapter-related protein complex 1 gamma 1
PDBTitle: ap1 clathrin adaptor core
|
| 11 | d1lrva_
|
|
 |
99.4 |
25 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Leucine-rich repeat variant |
| 12 | c2jkrL_
|
|
 |
99.2 |
10 |
PDB header:endocytosis
Chain: L: PDB Molecule:ap-2 complex subunit alpha-2;
PDBTitle: ap2 clathrin adaptor core with dileucine peptide rm(2 phosphos)qikrllse
|
| 13 | c3nmwA_
|
|
 |
99.1 |
13 |
PDB header:cell adhesion/cell cycle
Chain: A: PDB Molecule:apc variant protein;
PDBTitle: crytal structure of armadillo repeats domain of apc
|
| 14 | d1v18a1
|
|
 |
99.1 |
12 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 15 | d1xm9a1
|
|
 |
99.1 |
10 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Plakophilin 1 helical region |
| 16 | d1u6gc_
|
|
 |
99.0 |
14 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:HEAT repeat |
| 17 | d1b3ua_
|
|
 |
99.0 |
14 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:HEAT repeat |
| 18 | c1m5nS_
|
|
 |
99.0 |
10 |
PDB header:protein transport
Chain: S: PDB Molecule:importin beta-1 subunit;
PDBTitle: crystal structure of heat repeats (1-11) of importin b2 bound to the non-classical nls(67-94) of pthrp
|
| 19 | c3ifqB_
|
|
 |
98.9 |
14 |
PDB header:cell adhesion
Chain: B: PDB Molecule:plakoglobin;
PDBTitle: interction of plakoglobin and beta-catenin with desmosomal2 cadherins
|
| 20 | d1jdha_
|
|
 |
98.9 |
14 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 21 | c3nowA_ |
|
not modelled |
98.9 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:unc-45 protein, sd10334p;
PDBTitle: unc-45 from drosophila melanogaster
|
| 22 | c2z6hA_ |
|
not modelled |
98.9 |
15 |
PDB header:cell adhesion
Chain: A: PDB Molecule:catenin beta-1;
PDBTitle: crystal structure of beta-catenin armadillo repeat region2 and its c-terminal domain
|
| 23 | c4a0cB_ |
|
not modelled |
98.8 |
16 |
PDB header:cell cycle
Chain: B: PDB Molecule:cullin-associated nedd8-dissociated protein 1;
PDBTitle: structure of the cand1-cul4b-rbx1 complex
|
| 24 | d1lsha1 |
|
not modelled |
98.7 |
11 |
Fold:alpha-alpha superhelix
Superfamily:Lipovitellin-phosvitin complex, superhelical domain
Family:Lipovitellin-phosvitin complex, superhelical domain |
| 25 | c2z6gA_ |
|
not modelled |
98.7 |
13 |
PDB header:cell adhesion
Chain: A: PDB Molecule:b-catenin;
PDBTitle: crystal structure of a full-length zebrafish beta-catenin
|
| 26 | d1ibrb_ |
|
not modelled |
98.6 |
11 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 27 | d1ee4a_ |
|
not modelled |
98.6 |
17 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 28 | c3gq2B_ |
|
not modelled |
98.5 |
14 |
PDB header:transport protein
Chain: B: PDB Molecule:general vesicular transport factor p115;
PDBTitle: crystal structure of the dimer of the p115 tether globular head domain
|
| 29 | d1q1sc_ |
|
not modelled |
98.5 |
16 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 30 | c2w3cA_ |
|
not modelled |
98.5 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:general vesicular transport factor p115;
PDBTitle: globular head region of the human general vesicular2 transport factor p115
|
| 31 | c1pjmB_ |
|
not modelled |
98.4 |
16 |
PDB header:protein transport
Chain: B: PDB Molecule:importin alpha-2 subunit;
PDBTitle: mouse importin alpha-bipartite nls from human2 retinoblastoma protein complex
|
| 32 | c3bctA_ |
|
not modelled |
98.3 |
11 |
PDB header:armadillo repeat
Chain: A: PDB Molecule:beta-catenin;
PDBTitle: the armadillo repeat region from murine beta-catenin
|
| 33 | c3t7uA_ |
|
not modelled |
98.2 |
14 |
PDB header:cell adhesion
Chain: A: PDB Molecule:adenomatous polyposis coli protein;
PDBTitle: a new crytal structure of apc-arm
|
| 34 | c2qnaA_ |
|
not modelled |
98.2 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:importin subunit beta-1;
PDBTitle: crystal structure of human importin-beta (127-876) in complex with the2 ibb-domain of snurportin1 (1-65)
|
| 35 | c2c1tA_ |
|
not modelled |
98.2 |
17 |
PDB header:protein transport/membrane protein
Chain: A: PDB Molecule:importin alpha subunit;
PDBTitle: structure of the kap60p:nup2 complex
|
| 36 | c2ot8B_ |
|
not modelled |
98.2 |
15 |
PDB header:transport protein
Chain: B: PDB Molecule:transportin-1;
PDBTitle: karyopherin beta2/transportin-hnrnpm nls complex
|
| 37 | c1xqrA_ |
|
not modelled |
98.1 |
13 |
PDB header:chaperone
Chain: A: PDB Molecule:hspbp1 protein;
PDBTitle: crystal structure of the hspbp1 core domain
|
| 38 | c2jdqB_ |
|
not modelled |
98.1 |
15 |
PDB header:protein transport
Chain: B: PDB Molecule:importin alpha-1 subunit;
PDBTitle: c-terminal domain of influenza a virus polymerase pb22 subunit in complex with human importin alpha5
|
| 39 | c1lshA_ |
|
not modelled |
98.1 |
10 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:lipovitellin (lv-1n, lv-1c);
PDBTitle: lipid-protein interactions in lipovitellin
|
| 40 | d1xqra1 |
|
not modelled |
98.1 |
13 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:HspBP1 domain |
| 41 | c3nmzB_ |
|
not modelled |
98.0 |
10 |
PDB header:cell adhesion/cell cycle
Chain: B: PDB Molecule:apc variant protein;
PDBTitle: crytal structure of apc complexed with asef
|
| 42 | c2of3A_ |
|
not modelled |
98.0 |
13 |
PDB header:structural protein, cell cycle
Chain: A: PDB Molecule:zyg-9;
PDBTitle: tog domain structure from c.elegans zyg9
|
| 43 | c2qk2A_ |
|
not modelled |
98.0 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:lp04448p;
PDBTitle: structural basis of microtubule plus end tracking by xmap215, clip-1702 and eb1
|
| 44 | c2qk1A_ |
|
not modelled |
97.8 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:protein stu2;
PDBTitle: structural basis of microtubule plus end tracking by2 xmap215, clip-170 and eb1
|
| 45 | c1wa5B_ |
|
not modelled |
97.5 |
14 |
PDB header:nuclear transport
Chain: B: PDB Molecule:importin alpha subunit;
PDBTitle: crystal structure of the exportin cse1p complexed with its2 cargo (kap60p) and rangtp
|
| 46 | d1wa5b_ |
|
not modelled |
97.5 |
14 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 47 | d1qbkb_ |
|
not modelled |
97.3 |
12 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 48 | c3oqsA_ |
|
not modelled |
97.2 |
12 |
PDB header:protein transport
Chain: A: PDB Molecule:importin subunit alpha-2;
PDBTitle: crystal structure of importin-alpha bound to a clic4 nls peptide
|
| 49 | c3oc3B_ |
|
not modelled |
97.0 |
12 |
PDB header:hydrolase/transcription
Chain: B: PDB Molecule:helicase mot1;
PDBTitle: crystal structure of the mot1 n-terminal domain in complex with tbp
|
| 50 | d1qgra_ |
|
not modelled |
94.3 |
11 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 51 | d2bpta1 |
|
not modelled |
94.2 |
11 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 52 | d2i9ca1 |
|
not modelled |
91.5 |
16 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:RPA1889-like |
| 53 | c2wxoA_ |
|
not modelled |
90.2 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
PDBTitle: the crystal structure of the murine class ia pi 3-kinase2 p110delta in complex with as5.
|
| 54 | c3opbA_ |
|
not modelled |
87.6 |
12 |
PDB header:protein binding
Chain: A: PDB Molecule:swi5-dependent ho expression protein 4;
PDBTitle: crystal structure of she4p
|
| 55 | c2y3aA_ |
|
not modelled |
85.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
PDBTitle: crystal structure of p110beta in complex with icsh2 of p85beta and2 the drug gdc-0941
|
| 56 | c2rd0A_ |
|
not modelled |
84.9 |
12 |
PDB header:transferase/oncoprotein
Chain: A: PDB Molecule:phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
PDBTitle: structure of a human p110alpha/p85alpha complex
|
| 57 | d1ho8a_ |
|
not modelled |
83.3 |
7 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Regulatory subunit H of the V-type ATPase |
| 58 | d1e7ua1 |
|
not modelled |
82.8 |
17 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Phoshoinositide 3-kinase (PI3K) helical domain |
| 59 | c1e8zA_ |
|
not modelled |
80.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol 3-kinase catalytic subunit;
PDBTitle: structure determinants of phosphoinositide 3-kinase2 inhibition by wortmannin, ly294002, quercetin, myricetin3 and staurosporine
|
| 60 | c2l1lB_ |
|
not modelled |
80.6 |
14 |
PDB header:nuclear protein
Chain: B: PDB Molecule:exportin-1;
PDBTitle: nmr solution structure of the phi0 pki nes peptide in complex with2 crm1-rangtp
|
| 61 | c3o2qD_ |
|
not modelled |
76.8 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:symplekin;
PDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
|
| 62 | c3odsA_ |
|
not modelled |
75.9 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:symplekin;
PDBTitle: crystal structure of the k185a mutant of the n-terminal domain of2 human symplekin
|
| 63 | c3o2tA_ |
|
not modelled |
75.4 |
12 |
PDB header:protein binding
Chain: A: PDB Molecule:symplekin;
PDBTitle: crystal structure of the n-terminal domain of human symplekin
|
| 64 | c3odrA_ |
|
not modelled |
62.3 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:symplekin;
PDBTitle: crystal structure of the n-terminal domain of human symplekin
|
| 65 | d2b6ca1 |
|
not modelled |
59.0 |
12 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:BC3264-like |
| 66 | d1wa5c_ |
|
not modelled |
51.7 |
12 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Armadillo repeat |
| 67 | c2iw3B_ |
|
not modelled |
47.6 |
10 |
PDB header:translation
Chain: B: PDB Molecule:elongation factor 3a;
PDBTitle: elongation factor 3 in complex with adp
|
| 68 | c1vsy5_ |
|
not modelled |
44.9 |
17 |
PDB header:hydrolase
Chain: 5: PDB Molecule:proteasome activator blm10;
PDBTitle: proteasome activator complex
|
| 69 | d1z3xa1 |
|
not modelled |
38.3 |
18 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:GUN4-associated domain |
| 70 | c3gs3A_ |
|
not modelled |
34.8 |
15 |
PDB header:transcription, protein binding
Chain: A: PDB Molecule:symplekin;
PDBTitle: structure of the n-terminal heat domain of symplekin from d.2 melanogaster
|
| 71 | d1t06a_ |
|
not modelled |
33.4 |
9 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:BC3264-like |
| 72 | c2x6kB_ |
|
not modelled |
33.3 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:phosphotidylinositol 3 kinase 59f;
PDBTitle: the crystal structure of the drosophila class iii pi3-kinase2 vps34 in complex with pi-103
|
| 73 | d1l5ja1 |
|
not modelled |
28.1 |
24 |
Fold:alpha-alpha superhelix
Superfamily:Aconitase B, N-terminal domain
Family:Aconitase B, N-terminal domain |
| 74 | c3ls8A_ |
|
not modelled |
24.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol 3-kinase catalytic subunit
PDBTitle: crystal structure of human pik3c3 in complex with 3-[4-(4-2 morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol
|
| 75 | c3m1iC_ |
|
not modelled |
22.2 |
15 |
PDB header:protein transport
Chain: C: PDB Molecule:exportin-1;
PDBTitle: crystal structure of yeast crm1 (xpo1p) in complex with yeast ranbp12 (yrb1p) and yeast rangtp (gsp1pgtp)
|
| 76 | d2p7vb1 |
|
not modelled |
16.7 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 77 | c1dvpA_ |
|
not modelled |
16.2 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine
PDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
|
| 78 | c2l7kA_ |
|
not modelled |
12.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of protein cd1104.2 from clostridium difficile,2 northeast structural genomics consortium target cfr130
|
| 79 | c3zyqA_ |
|
not modelled |
12.8 |
13 |
PDB header:signaling
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine kinase
PDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
|
| 80 | c3ibvB_ |
|
not modelled |
10.6 |
12 |
PDB header:rna binding protein
Chain: B: PDB Molecule:exportin-t;
PDBTitle: karyopherin cytosolic state
|
| 81 | d1upka_ |
|
not modelled |
10.5 |
19 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Mo25 protein |
| 82 | c3gjxD_ |
|
not modelled |
9.9 |
21 |
PDB header:protein transport
Chain: D: PDB Molecule:exportin-1;
PDBTitle: crystal structure of the nuclear export complex crm1-2 snurportin1-rangtp
|
| 83 | d1x3zb1 |
|
not modelled |
9.9 |
12 |
Fold:XPC-binding domain
Superfamily:XPC-binding domain
Family:XPC-binding domain |
| 84 | c1x4qA_ |
|
not modelled |
7.3 |
11 |
PDB header:rna binding protein
Chain: A: PDB Molecule:u4/u6 small nuclear ribonucleoprotein prp3;
PDBTitle: solution structure of pwi domain in u4/u6 small nuclear2 ribonucleoprotein prp3(hprp3)
|
| 85 | c2qsgX_ |
|
not modelled |
6.6 |
12 |
PDB header:dna binding protein/dna
Chain: X: PDB Molecule:uv excision repair protein rad23;
PDBTitle: crystal structure of rad4-rad23 bound to a uv-damaged dna
|
| 86 | c2x19B_ |
|
not modelled |
6.5 |
11 |
PDB header:nuclear transport
Chain: B: PDB Molecule:importin-13;
PDBTitle: crystal structure of importin13 - rangtp complex
|
| 87 | c1l5jB_ |
|
not modelled |
6.1 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:aconitate hydratase 2;
PDBTitle: crystal structure of e. coli aconitase b.
|
| 88 | c3bvsA_ |
|
not modelled |
5.9 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkylpurine dna glycosylase alkd;
PDBTitle: crystal structure of bacillus cereus alkylpurine dna glycosylase alkd
|
| 89 | c1x5bA_ |
|
not modelled |
5.9 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:signal transducing adaptor molecule 2;
PDBTitle: the solution structure of the vhs domain of human signal2 transducing adaptor molecule 2
|
| 90 | d1dvpa1 |
|
not modelled |
5.5 |
10 |
Fold:alpha-alpha superhelix
Superfamily:ENTH/VHS domain
Family:VHS domain |
| 91 | d1bvsa1 |
|
not modelled |
5.4 |
23 |
Fold:RuvA C-terminal domain-like
Superfamily:DNA helicase RuvA subunit, C-terminal domain
Family:DNA helicase RuvA subunit, C-terminal domain |
| 92 | d1ttya_ |
|
not modelled |
5.1 |
8 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |