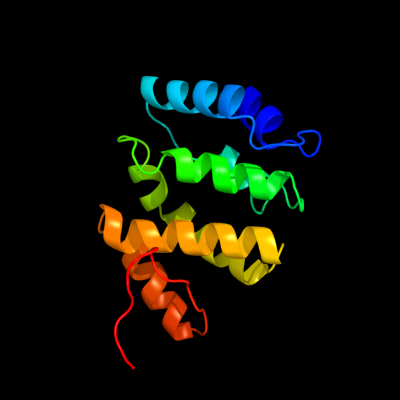
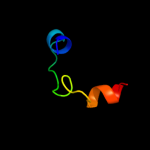
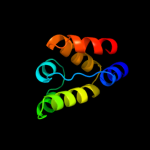
1 c1x5bA_
75.9
10
PDB header: protein bindingChain: A: PDB Molecule: signal transducing adaptor molecule 2;PDBTitle: the solution structure of the vhs domain of human signal2 transducing adaptor molecule 2
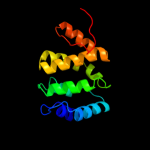
2 d1dvpa1
61.5
9
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain3 c3zyqA_
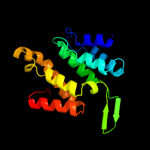
47.8
17
PDB header: signalingChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
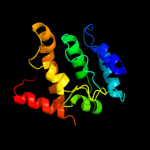
4 d1elka_
42.7
10
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain5 d2ezha_
42.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain6 c2w36B_
40.1
20
PDB header: hydrolaseChain: B: PDB Molecule: endonuclease v;PDBTitle: structures of endonuclease v with dna reveal initiation of2 deaminated adenine repair
7 c3i26C_
39.9
22
PDB header: hydrolaseChain: C: PDB Molecule: hemagglutinin-esterase;PDBTitle: structure of bovine torovirus hemagglutinin-esterase
8 d1ujka_
37.9
12
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain9 c3onlB_
37.1
14
PDB header: protein transportChain: B: PDB Molecule: epsin-3;PDBTitle: yeast ent3_enth-vti1p_habc complex structure
10 c3gocB_
36.9
33
PDB header: hydrolaseChain: B: PDB Molecule: endonuclease v;PDBTitle: crystal structure of the endonuclease v (sav1684) from streptomyces2 avermitilis. northeast structural genomics consortium target svr196
11 c3ga2A_
36.9
18
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease v;PDBTitle: crystal structure of the endonuclease_v (bsu36170) from2 bacillus subtilis, northeast structural genomics3 consortium target sr624
12 d1oz9a_
34.1
50
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase13 d2ezia_
33.5
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain14 c2r18A_
31.5
11
PDB header: viral proteinChain: A: PDB Molecule: capsid assembly protein vp3;PDBTitle: structural insights into the multifunctional protein vp3 of2 birnaviruses
15 d1xm5a_
31.1
42
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase16 d1tvia_
30.3
42
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase17 d1dp7p_
29.3
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: P4 origin-binding domain-like18 d1cy5a_
28.9
7
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD19 c1xaxA_
27.1
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0054 protein hi0004;PDBTitle: nmr structure of hi0004, a putative essential gene product2 from haemophilus influenzae
20 c3sqgE_
23.1
27
PDB header: transferaseChain: E: PDB Molecule: methyl-coenzyme m reductase, beta subunit;PDBTitle: crystal structure of a methyl-coenzyme m reductase purified from black2 sea mats
21 d1t1ua2
not modelled
22.0
19
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase22 c2eqoA_
not modelled
21.9
24
PDB header: transcriptionChain: A: PDB Molecule: tnf receptor-associated factor 3-interactingPDBTitle: solution structure of the stn_traf3ip1_nd domain of2 interleukin 13 receptor alpha 1-binding protein-1 [homo3 sapiens]
23 d1dmga_
not modelled
21.2
14
Fold: Ribosomal protein L4Superfamily: Ribosomal protein L4Family: Ribosomal protein L424 c3pvlA_
not modelled
20.5
14
PDB header: motor protein/protein transportChain: A: PDB Molecule: myosin viia isoform 1;PDBTitle: structure of myosin viia myth4-ferm-sh3 in complex with the cen1 of2 sans
25 c3ghgK_
not modelled
20.0
31
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
26 d2opla1
not modelled
19.5
19
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins27 d1ndba2
not modelled
19.3
20
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase28 c1h0mD_
not modelled
19.2
10
PDB header: transcription/dnaChain: D: PDB Molecule: transcriptional activator protein trar;PDBTitle: three-dimensional structure of the quorum sensing protein2 trar bound to its autoinducer and to its target dna
29 c1zljE_
not modelled
19.0
7
PDB header: transcriptionChain: E: PDB Molecule: dormancy survival regulator;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
30 c1hbmE_
not modelled
18.0
33
PDB header: methanogenesisChain: E: PDB Molecule: methyl-coenzyme m reductase i beta subunit;PDBTitle: methyl-coenzyme m reductase enzyme product complex
31 c3edeB_
not modelled
18.0
14
PDB header: hydrolaseChain: B: PDB Molecule: cyclomaltodextrinase;PDBTitle: structural base for cyclodextrin hydrolysis
32 d1a04a1
not modelled
17.9
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)33 d1h3ga3
not modelled
17.8
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain34 d1hbnb1
not modelled
17.7
33
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain35 c2vgqA_
not modelled
17.3
19
PDB header: immune system/transportChain: A: PDB Molecule: maltose-binding periplasmic protein,PDBTitle: crystal structure of human ips-1 card
36 c1deqO_
not modelled
17.2
33
PDB header: PDB COMPND: 37 d1nm8a2
not modelled
17.0
20
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase38 d1fs1b2
not modelled
16.6
33
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain39 c2gdvA_
not modelled
16.1
5
PDB header: transferaseChain: A: PDB Molecule: sucrose phosphorylase;PDBTitle: sucrose phosphorylase from bifidobacterium adolescentis2 reacted with sucrose
40 d1puza_
not modelled
15.7
15
Fold: YgfY-likeSuperfamily: YgfY-likeFamily: YgfY-like41 d1or7a1
not modelled
15.4
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain42 c2vdcF_
not modelled
15.0
14
PDB header: oxidoreductaseChain: F: PDB Molecule: glutamate synthase [nadph] large chain;PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
43 d1nexa2
not modelled
14.9
33
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain44 d2i3oa1
not modelled
14.5
10
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Gamma-glutamyltranspeptidase-like45 c2w31A_
not modelled
13.9
7
PDB header: oxygen transportChain: A: PDB Molecule: globin;PDBTitle: globin domain of geobacter sulfurreducens globin-coupled2 sensor
46 c1e6yE_
not modelled
13.9
33
PDB header: oxidoreductaseChain: E: PDB Molecule: methyl-coenzyme m reductase i beta subunit;PDBTitle: methyl-coenzyme m reductase from methanosarcina barkeri
47 d1xl7a2
not modelled
13.8
21
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase48 d2hc5a1
not modelled
13.8
13
Fold: FlaG-likeSuperfamily: FlaG-likeFamily: FlaG-like49 c2rnqA_
not modelled
13.6
36
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation factor iie subunitPDBTitle: solution structure of the c-terminal acidic domain of tfiie2 alpha
50 c2jtxA_
not modelled
13.6
36
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation factor iie subunitPDBTitle: nmr structure of the tfiie-alpha carboxyl terminus
51 c1dvpA_
not modelled
13.3
13
PDB header: transferaseChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosinePDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
52 c2lcvA_
not modelled
13.1
17
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional repressor cytr;PDBTitle: structure of the cytidine repressor dna-binding domain; an alternate2 calculation
53 d1e6yb1
not modelled
13.1
33
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain54 c3g5bA_
not modelled
13.0
16
PDB header: apoptosisChain: A: PDB Molecule: netrin receptor unc5b;PDBTitle: the structure of unc5b cytoplasmic domain
55 c3jroC_
not modelled
13.0
19
PDB header: transport protein, structural proteinChain: C: PDB Molecule: nucleoporin nup84;PDBTitle: nup84-nup145c-sec13 edge element of the npc lattice
56 c1xrxD_
not modelled
12.9
29
PDB header: replication inhibitorChain: D: PDB Molecule: seqa protein;PDBTitle: crystal structure of a dna-binding protein
57 d1xrxa1
not modelled
12.9
29
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: SeqA N-terminal domain-like58 c3qp5C_
not modelled
12.7
12
PDB header: transcriptionChain: C: PDB Molecule: cvir transcriptional regulator;PDBTitle: crystal structure of cvir bound to antagonist chlorolactone (cl)
59 d1e6vb1
not modelled
12.6
33
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain60 d1v74b_
not modelled
12.3
25
Fold: Four-helical up-and-down bundleSuperfamily: Colicin D immunity proteinFamily: Colicin D immunity protein61 d1hw6a_
not modelled
12.1
15
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)62 d1l3la1
not modelled
12.1
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)63 c2krfB_
not modelled
12.0
8
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein coma;PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
64 d1fsea_
not modelled
11.9
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)65 c3lvzA_
not modelled
11.9
12
PDB header: hydrolaseChain: A: PDB Molecule: blr6230 protein;PDBTitle: new refinement of the crystal structure of bjp-1, a subclass b32 metallo-beta-lactamase of bradyrhizobium japonicum
66 c3sztB_
not modelled
11.8
13
PDB header: transcriptionChain: B: PDB Molecule: quorum-sensing control repressor;PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
67 c2jq6A_
not modelled
11.3
17
PDB header: metal binding proteinChain: A: PDB Molecule: eh domain-containing protein 1;PDBTitle: structure of eh-domain of ehd1
68 d1q5ma_
not modelled
11.3
14
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)69 c2jpcA_
not modelled
11.1
10
PDB header: dna binding proteinChain: A: PDB Molecule: ssrb;PDBTitle: ssrb dna binding protein
70 c2hpcF_
not modelled
11.1
38
PDB header: blood clottingChain: F: PDB Molecule: fibrinogen, gamma polypeptide;PDBTitle: crystal structure of fragment d from human fibrinogen complexed with2 gly-pro-arg-pro-amide.
71 d1gvia3
not modelled
11.0
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain72 c3fiaA_
not modelled
10.9
13
PDB header: protein bindingChain: A: PDB Molecule: intersectin-1;PDBTitle: crystal structure of the eh 1 domain from human intersectin-2 1 protein. northeast structural genomics consortium target3 hr3646e.
73 c1qhoA_
not modelled
10.9
13
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: five-domain alpha-amylase from bacillus stearothermophilus,2 maltose/acarbose complex
74 d2pbea1
not modelled
10.8
18
Fold: PAP/OAS1 substrate-binding domainSuperfamily: PAP/OAS1 substrate-binding domainFamily: AadK C-terminal domain-like75 c3kblA_
not modelled
10.8
38
PDB header: protein bindingChain: A: PDB Molecule: female germline-specific tumor suppressor gld-1;PDBTitle: crystal structure of the gld-1 homodimerization domain from2 caenorhabditis elegans n169a mutant at 2.28 a resolution
76 d2ve8a1
not modelled
10.8
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FtsK C-terminal domain-like77 c2qy7A_
not modelled
10.6
18
PDB header: protein bindingChain: A: PDB Molecule: clathrin interactor 1;PDBTitle: crystal structure of human epsinr enth domain
78 d1q7ha2
not modelled
10.6
46
Fold: Cystatin-likeSuperfamily: Pre-PUA domainFamily: Hypothetical protein Ta1423, N-terminal domain79 d1tuka1
not modelled
10.6
27
Fold: Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albuminSuperfamily: Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albuminFamily: Plant lipid-transfer and hydrophobic proteins80 d1c01a_
not modelled
10.5
27
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Plant antimicrobial protein MIAMP181 d1nn7a_
not modelled
10.3
14
Fold: POZ domainSuperfamily: POZ domainFamily: Tetramerization domain of potassium channels82 d1j20a2
not modelled
10.3
17
Fold: Argininosuccinate synthetase, C-terminal domainSuperfamily: Argininosuccinate synthetase, C-terminal domainFamily: Argininosuccinate synthetase, C-terminal domain83 d1s1ga_
not modelled
10.3
14
Fold: POZ domainSuperfamily: POZ domainFamily: Tetramerization domain of potassium channels84 d2esha1
not modelled
10.3
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PadR-like85 c3bvhC_
not modelled
10.2
31
PDB header: blood clottingChain: C: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of recombinant gammad364a fibrinogen fragment d with2 the peptide ligand gly-pro-arg-pro-amide
86 c2kmgA_
not modelled
10.2
26
PDB header: gene regulationChain: A: PDB Molecule: klca;PDBTitle: the structure of the klca and ardb proteins show a novel2 fold and antirestriction activity against type i dna3 restriction systems in vivo but not in vitro
87 c2rnjA_
not modelled
10.2
15
PDB header: transcriptionChain: A: PDB Molecule: response regulator protein vrar;PDBTitle: nmr structure of the s. aureus vrar dna binding domain
88 d1ofda3
not modelled
10.1
21
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases89 c1nexC_
not modelled
10.0
36
PDB header: ligase, cell cycleChain: C: PDB Molecule: centromere dna-binding protein complex cbf3PDBTitle: crystal structure of scskp1-sccdc4-cpd peptide complex
90 c2q0oA_
not modelled
10.0
2
PDB header: transcriptionChain: A: PDB Molecule: probable transcriptional activator protein trar;PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
91 d2c9wc1
not modelled
10.0
36
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain92 c1lm1A_
not modelled
9.9
18
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin-dependent glutamate synthase;PDBTitle: structural studies on the synchronization of catalytic centers in2 glutamate synthase: native enzyme
93 d1ug7a_
not modelled
9.8
16
Fold: Four-helical up-and-down bundleSuperfamily: Domain from hypothetical 2610208m17rik proteinFamily: Domain from hypothetical 2610208m17rik protein94 c3k8kB_
not modelled
9.8
21
PDB header: membrane proteinChain: B: PDB Molecule: alpha-amylase, susg;PDBTitle: crystal structure of susg
95 d1ht6a2
not modelled
9.8
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain96 c2cblA_
not modelled
9.8
15
PDB header: complex (proto-oncogene/peptide)Chain: A: PDB Molecule: proto-oncogene cbl;PDBTitle: n-terminal domain of cbl in complex with its binding site2 on zap-70
97 d1inza_
not modelled
9.8
13
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: ENTH domain98 d1ea9c3
not modelled
9.8
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain99 c3excX_
not modelled
9.7
15
PDB header: hydrolaseChain: X: PDB Molecule: uncharacterized protein;PDBTitle: structure of the rna'se sso8090 from sulfolobus solfataricus