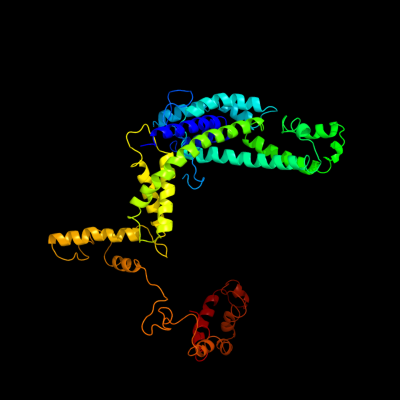
| 1 | c3iydF_
|
|
 |
100.0 |
96 |
PDB header:transcription/dna
Chain: F: PDB Molecule:rna polymerase sigma factor rpod;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
|
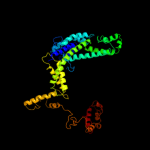
| 2 | c2a6eF_
|
|
 |
100.0 |
59 |
PDB header:transferase
Chain: F: PDB Molecule:rna polymerase sigma factor rpod;
PDBTitle: crystal structure of the t. thermophilus rna polymerase2 holoenzyme
|
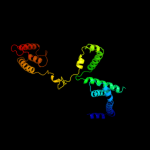
| 3 | c1l9uH_
|
|
 |
100.0 |
56 |
PDB header:transcription
Chain: H: PDB Molecule:sigma factor siga;
PDBTitle: thermus aquaticus rna polymerase holoenzyme at 4 a2 resolution
|
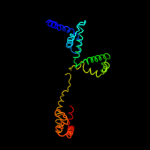
| 4 | c1ku2A_
|
|
 |
100.0 |
60 |
PDB header:transcription
Chain: A: PDB Molecule:sigma factor siga;
PDBTitle: crystal structure of thermus aquaticus rna polymerase sigma2 subunit fragment containing regions 1.2 to 3.1
|
| 5 | d1siga_
|
|
 |
100.0 |
86 |
Fold:Sigma2 domain of RNA polymerase sigma factors
Superfamily:Sigma2 domain of RNA polymerase sigma factors
Family:Sigma2 domain of RNA polymerase sigma factors |
| 6 | c1rp3G_
|
|
 |
100.0 |
25 |
PDB header:transcription
Chain: G: PDB Molecule:rna polymerase sigma factor sigma-28 (flia);
PDBTitle: cocrystal structure of the flagellar sigma/anti-sigma2 complex, sigma-28/flgm
|
| 7 | d1ku2a2
|
|
 |
100.0 |
60 |
Fold:Sigma2 domain of RNA polymerase sigma factors
Superfamily:Sigma2 domain of RNA polymerase sigma factors
Family:Sigma2 domain of RNA polymerase sigma factors |
| 8 | d1smyf3
|
|
 |
100.0 |
60 |
Fold:Sigma2 domain of RNA polymerase sigma factors
Superfamily:Sigma2 domain of RNA polymerase sigma factors
Family:Sigma2 domain of RNA polymerase sigma factors |
| 9 | d1rp3a3
|
|
 |
99.8 |
28 |
Fold:Sigma2 domain of RNA polymerase sigma factors
Superfamily:Sigma2 domain of RNA polymerase sigma factors
Family:Sigma2 domain of RNA polymerase sigma factors |
| 10 | c3t72o_
|
|
 |
99.7 |
95 |
PDB header:transcription/dna
Chain: O: PDB Molecule:pho box dna (strand 1);
PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
|
| 11 | d1smyf2
|
|
 |
99.7 |
56 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 12 | d1ku7a_
|
|
 |
99.6 |
56 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 13 | d2p7vb1
|
|
 |
99.6 |
100 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 14 | d1ttya_
|
|
 |
99.5 |
62 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 15 | c1or7A_
|
|
 |
99.4 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma-e factor;
PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
|
| 16 | c3mzyA_
|
|
 |
99.4 |
31 |
PDB header:rna binding protein
Chain: A: PDB Molecule:rna polymerase sigma-h factor;
PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
|
| 17 | d1ku3a_
|
|
 |
99.4 |
56 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 18 | c2q1zA_
|
|
 |
99.3 |
20 |
PDB header:transcription
Chain: A: PDB Molecule:rpoe, ecf sige;
PDBTitle: crystal structure of rhodobacter sphaeroides sige in complex with the2 anti-sigma chrr
|
| 19 | d1ku2a1
|
|
 |
99.2 |
59 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
| 20 | d1l0oc_
|
|
 |
98.9 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 21 | c1l0oC_ |
|
not modelled |
98.9 |
18 |
PDB header:protein binding
Chain: C: PDB Molecule:sigma factor;
PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
|
| 22 | d1rp3a2 |
|
not modelled |
98.9 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 23 | c3hugA_ |
|
not modelled |
98.8 |
19 |
PDB header:transcription/membrane protein
Chain: A: PDB Molecule:rna polymerase sigma factor;
PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
|
| 24 | d1rp3a1 |
|
not modelled |
98.7 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
| 25 | d1or7a1 |
|
not modelled |
98.3 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 26 | c2o8xA_ |
|
not modelled |
98.2 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:probable rna polymerase sigma-c factor;
PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
|
| 27 | d1xsva_ |
|
not modelled |
98.2 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:YlxM/p13-like |
| 28 | d1s7oa_ |
|
not modelled |
98.1 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:YlxM/p13-like |
| 29 | c1c1gA_ |
|
not modelled |
98.0 |
8 |
PDB header:contractile protein
Chain: A: PDB Molecule:tropomyosin;
PDBTitle: crystal structure of tropomyosin at 7 angstroms resolution2 in the spermine-induced crystal form
|
| 30 | c1ciiA_
|
|
 |
97.9 |
12 |
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
|
| 31 | d1or7b2 |
|
not modelled |
97.8 |
23 |
Fold:Sigma2 domain of RNA polymerase sigma factors
Superfamily:Sigma2 domain of RNA polymerase sigma factors
Family:Sigma2 domain of RNA polymerase sigma factors |
| 32 | d1or7a2 |
|
not modelled |
97.8 |
21 |
Fold:Sigma2 domain of RNA polymerase sigma factors
Superfamily:Sigma2 domain of RNA polymerase sigma factors
Family:Sigma2 domain of RNA polymerase sigma factors |
| 33 | d1yioa1 |
|
not modelled |
97.7 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 34 | c3ojaB_ |
|
not modelled |
97.4 |
13 |
PDB header:protein binding
Chain: B: PDB Molecule:anopheles plasmodium-responsive leucine-rich repeat protein
PDBTitle: crystal structure of lrim1/apl1c complex
|
| 35 | c1x3uA_ |
|
not modelled |
97.2 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein fixj;
PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
|
| 36 | c2k6xA_
|
|
 |
97.1 |
28 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpod;
PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
|
| 37 | c1yvlB_ |
|
not modelled |
97.0 |
7 |
PDB header:signaling protein
Chain: B: PDB Molecule:signal transducer and activator of transcription
PDBTitle: structure of unphosphorylated stat1
|
| 38 | c1zljE_ |
|
not modelled |
97.0 |
18 |
PDB header:transcription
Chain: E: PDB Molecule:dormancy survival regulator;
PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
|
| 39 | c3qp5C_ |
|
not modelled |
97.0 |
24 |
PDB header:transcription
Chain: C: PDB Molecule:cvir transcriptional regulator;
PDBTitle: crystal structure of cvir bound to antagonist chlorolactone (cl)
|
| 40 | d1a04a1 |
|
not modelled |
97.0 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 41 | c2o7gA_ |
|
not modelled |
96.9 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:probable rna polymerase sigma-c factor;
PDBTitle: crystal structure of the pribnow box recognition region of sigc from2 mycobacterium tuberculosis
|
| 42 | c2rnjA_ |
|
not modelled |
96.9 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator protein vrar;
PDBTitle: nmr structure of the s. aureus vrar dna binding domain
|
| 43 | d1fsea_ |
|
not modelled |
96.8 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 44 | c2krfB_ |
|
not modelled |
96.8 |
22 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulatory protein coma;
PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
|
| 45 | c2q0oA_ |
|
not modelled |
96.8 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:probable transcriptional activator protein trar;
PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
|
| 46 | d1l3la1 |
|
not modelled |
96.7 |
27 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 47 | c3sztB_ |
|
not modelled |
96.7 |
26 |
PDB header:transcription
Chain: B: PDB Molecule:quorum-sensing control repressor;
PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
|
| 48 | c1h0mD_ |
|
not modelled |
96.7 |
26 |
PDB header:transcription/dna
Chain: D: PDB Molecule:transcriptional activator protein trar;
PDBTitle: three-dimensional structure of the quorum sensing protein2 trar bound to its autoinducer and to its target dna
|
| 49 | d1p4wa_ |
|
not modelled |
96.5 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 50 | c3cloC_ |
|
not modelled |
96.5 |
21 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator containing a2 luxr dna binding domain (np_811094.1) from bacteroides3 thetaiotaomicron vpi-5482 at 2.04 a resolution
|
| 51 | c2w48D_ |
|
not modelled |
96.0 |
15 |
PDB header:transcription
Chain: D: PDB Molecule:sorbitol operon regulator;
PDBTitle: crystal structure of the full-length sorbitol operon2 regulator sorc from klebsiella pneumoniae
|
| 52 | c3cwgA_ |
|
not modelled |
95.9 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:signal transducer and activator of transcription
PDBTitle: unphosphorylated mouse stat3 core fragment
|
| 53 | c3n0rA_ |
|
not modelled |
95.5 |
19 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator;
PDBTitle: structure of the phyr stress response regulator at 1.25 angstrom2 resolution
|
| 54 | d1h3la_ |
|
not modelled |
95.5 |
16 |
Fold:Sigma2 domain of RNA polymerase sigma factors
Superfamily:Sigma2 domain of RNA polymerase sigma factors
Family:Sigma2 domain of RNA polymerase sigma factors |
| 55 | c3klnC_ |
|
not modelled |
95.4 |
17 |
PDB header:transcription
Chain: C: PDB Molecule:transcriptional regulator, luxr family;
PDBTitle: vibrio cholerae vpst
|
| 56 | c3c3wB_ |
|
not modelled |
95.4 |
20 |
PDB header:transcription
Chain: B: PDB Molecule:two component transcriptional regulatory protein devr;
PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
|
| 57 | c1bg1A_ |
|
not modelled |
95.4 |
8 |
PDB header:transcription/dna
Chain: A: PDB Molecule:protein (transcription factor stat3b);
PDBTitle: transcription factor stat3b/dna complex
|
| 58 | c2jpcA_ |
|
not modelled |
95.3 |
22 |
PDB header:dna binding protein
Chain: A: PDB Molecule:ssrb;
PDBTitle: ssrb dna binding protein
|
| 59 | c1ei3C_ |
|
not modelled |
94.9 |
4 |
PDB header:
PDB COMPND:
|
| 60 | c1bf5A_ |
|
not modelled |
94.6 |
7 |
PDB header:gene regulation/dna
Chain: A: PDB Molecule:signal transducer and activator of transcription
PDBTitle: tyrosine phosphorylated stat-1/dna complex
|
| 61 | c2efrB_ |
|
not modelled |
94.5 |
12 |
PDB header:contractile protein
Chain: B: PDB Molecule:general control protein gcn4 and tropomyosin 1 alpha chain;
PDBTitle: crystal structure of the c-terminal tropomyosin fragment with n- and2 c-terminal extensions of the leucine zipper at 1.8 angstroms3 resolution
|
| 62 | c1hciB_ |
|
not modelled |
94.3 |
9 |
PDB header:triple-helix coiled coil
Chain: B: PDB Molecule:alpha-actinin 2;
PDBTitle: crystal structure of the rod domain of alpha-actinin
|
| 63 | c1deqF_ |
|
not modelled |
94.3 |
9 |
PDB header:
PDB COMPND:
|
| 64 | c1zn2A_ |
|
not modelled |
94.1 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:response regulatory protein;
PDBTitle: low resolution structure of response regulator styr
|
| 65 | c1rnlA_ |
|
not modelled |
93.7 |
22 |
PDB header:signal transduction protein
Chain: A: PDB Molecule:nitrate/nitrite response regulator protein narl;
PDBTitle: the nitrate/nitrite response regulator protein narl from narl
|
| 66 | c3na7A_ |
|
not modelled |
93.6 |
9 |
PDB header:gene regulation, chaperone
Chain: A: PDB Molecule:hp0958;
PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
|
| 67 | c2oevA_ |
|
not modelled |
93.3 |
8 |
PDB header:protein transport
Chain: A: PDB Molecule:programmed cell death 6-interacting protein;
PDBTitle: crystal structure of alix/aip1
|
| 68 | d1jhfa1 |
|
not modelled |
92.9 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:LexA repressor, N-terminal DNA-binding domain |
| 69 | c1u78A_ |
|
not modelled |
92.9 |
22 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:transposable element tc3 transposase;
PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
|
| 70 | c2d3eD_ |
|
not modelled |
92.7 |
15 |
PDB header:contractile protein
Chain: D: PDB Molecule:general control protein gcn4 and tropomyosin 1
PDBTitle: crystal structure of the c-terminal fragment of rabbit2 skeletal alpha-tropomyosin
|
| 71 | c1jchC_ |
|
not modelled |
92.3 |
11 |
PDB header:ribosome inhibitor, hydrolase
Chain: C: PDB Molecule:colicin e3;
PDBTitle: crystal structure of colicin e3 in complex with its immunity protein
|
| 72 | d2o38a1 |
|
not modelled |
92.0 |
11 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:NE1354 |
| 73 | c2o38A_ |
|
not modelled |
92.0 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: putative xre family transcriptional regulator
|
| 74 | c2v71A_ |
|
not modelled |
91.3 |
10 |
PDB header:nuclear protein
Chain: A: PDB Molecule:nuclear distribution protein nude-like 1;
PDBTitle: coiled-coil region of nudel
|
| 75 | c3f2hA_ |
|
not modelled |
91.2 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:alkylmercury lyase;
PDBTitle: crystal structure of the mercury-bound form of merb mutant2 c160s, the organomercurial lyase involved in a bacterial3 mercury resistance system
|
| 76 | c3mkyP_ |
|
not modelled |
89.9 |
16 |
PDB header:dna binding protein/dna
Chain: P: PDB Molecule:protein sopb;
PDBTitle: structure of sopb(155-323)-18mer dna complex, i23 form
|
| 77 | d2a6ca1 |
|
not modelled |
89.9 |
15 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:NE1354 |
| 78 | d1trra_ |
|
not modelled |
89.8 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Trp repressor, TrpR |
| 79 | c1deqO_ |
|
not modelled |
89.7 |
10 |
PDB header:
PDB COMPND:
|
| 80 | c3r0aB_ |
|
not modelled |
89.2 |
11 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator;
PDBTitle: possible transcriptional regulator from methanosarcina mazei go1 (gi2 21227196)
|
| 81 | c3ghgK_ |
|
not modelled |
88.8 |
4 |
PDB header:blood clotting
Chain: K: PDB Molecule:fibrinogen beta chain;
PDBTitle: crystal structure of human fibrinogen
|
| 82 | d2d1ha1 |
|
not modelled |
88.8 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:TrmB-like |
| 83 | c2x4hA_ |
|
not modelled |
88.7 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:hypothetical protein sso2273;
PDBTitle: crystal structure of the hypothetical protein sso2273 from2 sulfolobus solfataricus
|
| 84 | c3ojaA_ |
|
not modelled |
88.3 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:leucine-rich immune molecule 1;
PDBTitle: crystal structure of lrim1/apl1c complex
|
| 85 | c1ei3E_ |
|
not modelled |
88.0 |
5 |
PDB header:
PDB COMPND:
|
| 86 | c1deqD_ |
|
not modelled |
87.8 |
5 |
PDB header:
PDB COMPND:
|
| 87 | c3hnwB_ |
|
not modelled |
87.5 |
8 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
|
| 88 | c3fmyA_ |
|
not modelled |
87.4 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:hth-type transcriptional regulator mqsa
PDBTitle: structure of the c-terminal domain of the e. coli protein2 mqsa (ygit/b3021)
|
| 89 | d1vz0a1 |
|
not modelled |
87.0 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:KorB DNA-binding domain-like
Family:KorB DNA-binding domain-like |
| 90 | c2w7nA_ |
|
not modelled |
87.0 |
9 |
PDB header:transcription/dna
Chain: A: PDB Molecule:trfb transcriptional repressor protein;
PDBTitle: crystal structure of kora bound to operator dna: insight2 into repressor cooperation in rp4 gene regulation
|
| 91 | c2bnoA_ |
|
not modelled |
86.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:epoxidase;
PDBTitle: the structure of hydroxypropylphosphonic acid epoxidase2 from s. wedmorenis.
|
| 92 | c3mlfC_ |
|
not modelled |
86.7 |
26 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulator;
PDBTitle: putative transcriptional regulator from staphylococcus aureus.
|
| 93 | d1rioa_ |
|
not modelled |
86.6 |
8 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:Phage repressors |
| 94 | c3k2zA_ |
|
not modelled |
85.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:lexa repressor;
PDBTitle: crystal structure of a lexa protein from thermotoga maritima
|
| 95 | c1y9qA_ |
|
not modelled |
85.5 |
18 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, hth_3 family;
PDBTitle: crystal structure of hth_3 family transcriptional regulator2 from vibrio cholerae
|
| 96 | c3dtpA_ |
|
not modelled |
85.3 |
11 |
PDB header:contractile protein
Chain: A: PDB Molecule:myosin 2 heavy chain chimera of smooth and
PDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
|
| 97 | d2cfxa1 |
|
not modelled |
85.0 |
7 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Lrp/AsnC-like transcriptional regulator N-terminal domain |
| 98 | c2p6tH_ |
|
not modelled |
85.0 |
16 |
PDB header:transcription
Chain: H: PDB Molecule:transcriptional regulator, lrp/asnc family;
PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
|
| 99 | c3kxaD_ |
|
not modelled |
83.9 |
20 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of ngo0477 from neisseria gonorrhoeae
|
| 100 | d2cg4a1 |
|
not modelled |
83.7 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Lrp/AsnC-like transcriptional regulator N-terminal domain |
| 101 | d1biaa1 |
|
not modelled |
83.6 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Biotin repressor-like |
| 102 | c2l4aA_ |
|
not modelled |
83.4 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:leucine responsive regulatory protein;
PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
|
| 103 | d1nr3a_ |
|
not modelled |
83.4 |
19 |
Fold:DNA-binding protein Tfx
Superfamily:DNA-binding protein Tfx
Family:DNA-binding protein Tfx |
| 104 | d2b5aa1 |
|
not modelled |
83.3 |
18 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:SinR domain-like |
| 105 | d1i1ga1 |
|
not modelled |
83.1 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Lrp/AsnC-like transcriptional regulator N-terminal domain |
| 106 | c2it0A_ |
|
not modelled |
83.1 |
13 |
PDB header:transcription/dna
Chain: A: PDB Molecule:iron-dependent repressor ider;
PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
|
| 107 | c3nqoB_ |
|
not modelled |
83.0 |
13 |
PDB header:transcription
Chain: B: PDB Molecule:marr-family transcriptional regulator;
PDBTitle: crystal structure of a marr family transcriptional regulator (cd1569)2 from clostridium difficile 630 at 2.20 a resolution
|
| 108 | c2jvlA_ |
|
not modelled |
82.8 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:trmbf1;
PDBTitle: nmr structure of the c-terminal domain of mbf1 of trichoderma reesei
|
| 109 | c3trbA_ |
|
not modelled |
82.8 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:virulence-associated protein i;
PDBTitle: structure of an addiction module antidote protein of a higa (higa)2 family from coxiella burnetii
|
| 110 | d1r71a_ |
|
not modelled |
82.7 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:KorB DNA-binding domain-like
Family:KorB DNA-binding domain-like |
| 111 | d1y7ya1 |
|
not modelled |
82.6 |
12 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:SinR domain-like |
| 112 | c2ef8A_ |
|
not modelled |
82.6 |
15 |
PDB header:transcription regulator
Chain: A: PDB Molecule:putative transcription factor;
PDBTitle: crystal structure of c.ecot38is
|
| 113 | d1lmb3_ |
|
not modelled |
82.4 |
9 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:Phage repressors |
| 114 | c2vbzA_ |
|
not modelled |
82.0 |
14 |
PDB header:dna-binding protein
Chain: A: PDB Molecule:transcriptional regulatory protein;
PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
|
| 115 | d1jhga_ |
|
not modelled |
81.7 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Trp repressor, TrpR |
| 116 | c3f6wE_ |
|
not modelled |
81.6 |
15 |
PDB header:dna binding protein
Chain: E: PDB Molecule:xre-family like protein;
PDBTitle: xre-family like protein from pseudomonas syringae pv. tomato str.2 dc3000
|
| 117 | c1r71B_ |
|
not modelled |
81.6 |
21 |
PDB header:transcription/dna
Chain: B: PDB Molecule:transcriptional repressor protein korb;
PDBTitle: crystal structure of the dna binding domain of korb in2 complex with the operator dna
|
| 118 | c2gl2B_ |
|
not modelled |
81.5 |
11 |
PDB header:cell adhesion
Chain: B: PDB Molecule:adhesion a;
PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
|
| 119 | c2ppxA_ |
|
not modelled |
81.3 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu1735;
PDBTitle: crystal structure of a hth xre-family like protein from agrobacterium2 tumefaciens
|
| 120 | d2ppxa1 |
|
not modelled |
81.3 |
19 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:SinR domain-like |