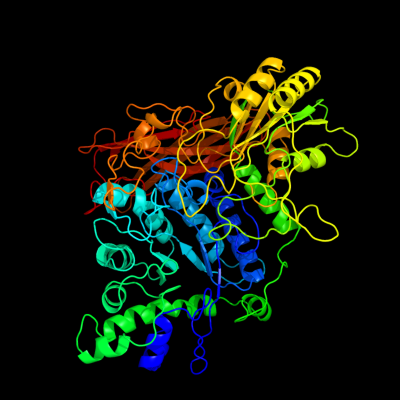
| 1 | c3ac0B_
|
|
 |
100.0 |
33 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase i;
PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
|
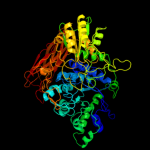
| 2 | c2x41A_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana2 in complex with glucose
|
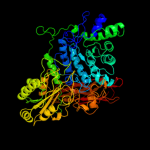
| 3 | c3f93D_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of exo-1,3/1,4-beta-glucanase (exop) from2 pseudoalteromonas sp. bb1
|
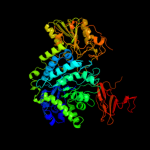
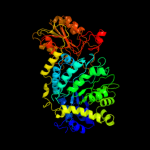
| 4 | c1ex1A_
|
|
 |
100.0 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (beta-d-glucan exohydrolase isoenzyme exo1);
PDBTitle: beta-d-glucan exohydrolase from barley
|
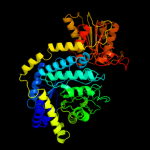
| 5 | c3bmxB_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
|
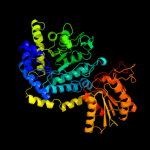
| 6 | c3lk6A_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
|
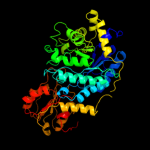
| 7 | c3sqlB_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3;
PDBTitle: crystal structure of glycoside hydrolase from synechococcus
|
| 8 | d1x38a1
|
|
 |
100.0 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:NagZ-like |
| 9 | c3tevA_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hyrolase, family 3;
PDBTitle: the crystal structure of glycosyl hydrolase from deinococcus2 radiodurans r1
|
| 10 | d1tr9a_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:NagZ-like |
| 11 | d1x38a2
|
|
 |
100.0 |
35 |
Fold:Flavodoxin-like
Superfamily:Beta-D-glucan exohydrolase, C-terminal domain
Family:Beta-D-glucan exohydrolase, C-terminal domain |
| 12 | c2kl6A_
|
|
 |
97.4 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the cardb domain of pf1109 from2 pyrococcus furiosus. northeast structural genomics3 consortium target pfr193a
|
| 13 | d1w8oa1
|
|
 |
97.3 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:E-set domains of sugar-utilizing enzymes |
| 14 | c2l0dA_
|
|
 |
97.1 |
14 |
PDB header:cell adhesion
Chain: A: PDB Molecule:cell surface protein;
PDBTitle: solution nmr structure of putative cell surface protein ma_4588 (272-2 376 domain) from methanosarcina acetivorans, northeast structural3 genomics consortium target mvr254a
|
| 15 | c2kutA_
|
|
 |
96.9 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of gmr58a from geobacter metallireducens.2 northeast structural genomics consortium target gmr58a
|
| 16 | d2vzsa2
|
|
 |
96.2 |
27 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 17 | c1l9mB_
|
|
 |
95.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:protein-glutamine glutamyltransferase e3;
PDBTitle: three-dimensional structure of the human transglutaminase 32 enzyme: binding of calcium ions change structure for3 activation
|
| 18 | d2q3za2
|
|
 |
95.8 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
| 19 | c2x3bB_
|
|
 |
95.3 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:toxic extracellular endopeptidase;
PDBTitle: asap1 inactive mutant e294a, an extracellular toxic zinc2 metalloendopeptidase
|
| 20 | d1ex0a2
|
|
 |
94.4 |
12 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
| 21 | d1g0da2 |
|
not modelled |
94.0 |
12 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
| 22 | c3isyA_ |
|
not modelled |
93.5 |
22 |
PDB header:protein binding
Chain: A: PDB Molecule:intracellular proteinase inhibitor;
PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
|
| 23 | d1vjja2 |
|
not modelled |
93.2 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
| 24 | c3rgbA_ |
|
not modelled |
90.6 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methane monooxygenase subunit b2;
PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
|
| 25 | d4ubpb_ |
|
not modelled |
89.1 |
22 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 26 | d2co7b1 |
|
not modelled |
88.8 |
18 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:PapD-like
Family:Pilus chaperone |
| 27 | c1yewI_ |
|
not modelled |
88.7 |
25 |
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
|
| 28 | c3fn9B_ |
|
not modelled |
88.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
| 29 | c2e6jA_ |
|
not modelled |
87.8 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hydin protein;
PDBTitle: solution structure of the c-terminal papd-like domain from2 human hydin protein
|
| 30 | d1ejxb_ |
|
not modelled |
87.6 |
24 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 31 | d1e9ya1 |
|
not modelled |
86.0 |
26 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 32 | c2cqtA_ |
|
not modelled |
86.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:cellobiose phosphorylase;
PDBTitle: crystal structure of cellvibrio gilvus cellobiose phosphorylase2 crystallized from sodium/potassium phosphate
|
| 33 | c1kv3F_ |
|
not modelled |
85.1 |
17 |
PDB header:transferase
Chain: F: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: human tissue transglutaminase in gdp bound form
|
| 34 | c3qgaD_ |
|
not modelled |
84.4 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:fusion of urease beta and gamma subunits;
PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
|
| 35 | c1g0dA_ |
|
not modelled |
83.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: crystal structure of red sea bream transglutaminase
|
| 36 | c1f13A_ |
|
not modelled |
83.5 |
12 |
PDB header:coagulation factor
Chain: A: PDB Molecule:cellular coagulation factor xiii zymogen;
PDBTitle: recombinant human cellular coagulation factor xiii
|
| 37 | d1v7wa2 |
|
not modelled |
81.8 |
15 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Glycosyltransferase family 36 N-terminal domain |
| 38 | c1iq8B_ |
|
not modelled |
80.6 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:archaeosine trna-guanine transglycosylase;
PDBTitle: crystal structure of archaeosine trna-guanine2 transglycosylase from pyrococcus horikoshii
|
| 39 | d1jz8a1 |
|
not modelled |
80.1 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 40 | c1e9zA_ |
|
not modelled |
80.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:urease subunit alpha;
PDBTitle: crystal structure of helicobacter pylori urease
|
| 41 | c2a74B_ |
|
not modelled |
79.5 |
14 |
PDB header:immune system
Chain: B: PDB Molecule:complement component c3c;
PDBTitle: human complement component c3c
|
| 42 | d2ccwa1 |
|
not modelled |
78.9 |
18 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 43 | c3rfrI_ |
|
not modelled |
78.5 |
18 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:pmob;
PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
|
| 44 | c3eoeC_ |
|
not modelled |
77.8 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007
|
| 45 | c3qisA_ |
|
not modelled |
77.0 |
20 |
PDB header:hydrolase/protein binding
Chain: A: PDB Molecule:inositol polyphosphate 5-phosphatase ocrl-1;
PDBTitle: recognition of the f&h motif by the lowe syndrome protein ocrl
|
| 46 | c2h47C_ |
|
not modelled |
76.6 |
26 |
PDB header:oxidoreductase/electron transport
Chain: C: PDB Molecule:azurin;
PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
|
| 47 | c3qbtH_ |
|
not modelled |
75.5 |
15 |
PDB header:protein transport/hydrolase
Chain: H: PDB Molecule:inositol polyphosphate 5-phosphatase ocrl-1;
PDBTitle: crystal structure of ocrl1 540-678 in complex with rab8a:gppnhp
|
| 48 | c2qsvA_ |
|
not modelled |
75.2 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function from porphyromonas2 gingivalis w83
|
| 49 | c1v7wA_ |
|
not modelled |
74.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:chitobiose phosphorylase;
PDBTitle: crystal structure of vibrio proteolyticus chitobiose phosphorylase in2 complex with glcnac
|
| 50 | c3hs0B_ |
|
not modelled |
74.6 |
13 |
PDB header:immune system
Chain: B: PDB Molecule:cobra venom factor;
PDBTitle: cobra venom factor (cvf) in complex with human factor b
|
| 51 | d2g50a2 |
|
not modelled |
74.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 52 | c5mdhB_ |
|
not modelled |
73.2 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of ternary complex of porcine cytoplasmic malate2 dehydrogenase alpha-ketomalonate and tnad at 2.4 angstroms resolution
|
| 53 | d1azca_ |
|
not modelled |
72.9 |
18 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 54 | d1civa1 |
|
not modelled |
72.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 55 | d7mdha1 |
|
not modelled |
72.5 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 56 | d1liua2 |
|
not modelled |
72.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 57 | c2co7B_ |
|
not modelled |
72.1 |
18 |
PDB header:fibril protein
Chain: B: PDB Molecule:putative fimbriae assembly chaperone;
PDBTitle: salmonella enterica safa pilin in complex with the safb2 chaperone (type ii)
|
| 58 | c3g6jB_ |
|
not modelled |
71.9 |
15 |
PDB header:immune system
Chain: B: PDB Molecule:complement c3 alpha chain;
PDBTitle: c3b in complex with a c3b specific fab
|
| 59 | c1mldA_ |
|
not modelled |
71.7 |
21 |
PDB header:oxidoreductase(nad(a)-choh(d))
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: refined structure of mitochondrial malate dehydrogenase2 from porcine heart and the consensus structure for3 dicarboxylic acid oxidoreductases
|
| 60 | d1y7ta1 |
|
not modelled |
71.6 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 61 | d5mdha1 |
|
not modelled |
71.5 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 62 | c1b8vA_ |
|
not modelled |
71.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (malate dehydrogenase);
PDBTitle: malate dehydrogenase from aquaspirillum arcticum
|
| 63 | c3fcsA_ |
|
not modelled |
69.7 |
15 |
PDB header:cell adhesion/blood clotting
Chain: A: PDB Molecule:integrin, alpha 2b;
PDBTitle: structure of complete ectodomain of integrin aiibb3
|
| 64 | d1b8pa1 |
|
not modelled |
69.1 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 65 | c3t07D_ |
|
not modelled |
68.8 |
23 |
PDB header:transferase/transferase inhibitor
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
|
| 66 | c1e0tD_ |
|
not modelled |
68.6 |
14 |
PDB header:phosphotransferase
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: r292d mutant of e. coli pyruvate kinase
|
| 67 | c2ys4A_ |
|
not modelled |
68.3 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hydrocephalus-inducing protein homolog;
PDBTitle: solution structure of the n-terminal papd-like domain of2 hydin protein from human
|
| 68 | c2a73B_ |
|
not modelled |
67.8 |
14 |
PDB header:immune system
Chain: B: PDB Molecule:complement c3;
PDBTitle: human complement component c3
|
| 69 | c7mdhA_ |
|
not modelled |
66.6 |
21 |
PDB header:chloroplastic malate dehydrogenase
Chain: A: PDB Molecule:protein (malate dehydrogenase);
PDBTitle: structural basis for light acitvation of a chloroplast enzyme. the2 structure of sorghum nadp-malate dehydrogenase in its oxidized form
|
| 70 | d1cc3a_ |
|
not modelled |
66.5 |
16 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 71 | c1sevA_ |
|
not modelled |
66.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase, glyoxysomal precursor;
PDBTitle: mature and translocatable forms of glyoxysomal malate2 dehydrogenase have different activities and stabilities3 but similar crystal structures
|
| 72 | c1smkD_ |
|
not modelled |
66.4 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase, glyoxysomal;
PDBTitle: mature and translocatable forms of glyoxysomal malate2 dehydrogenase have different activities and stabilities3 but similar crystal structures
|
| 73 | d1cuoa_ |
|
not modelled |
66.1 |
20 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 74 | d1llda1 |
|
not modelled |
65.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 75 | c2rdo7_ |
|
not modelled |
65.5 |
15 |
PDB header:ribosome
Chain: 7: PDB Molecule:elongation factor g;
PDBTitle: 50s subunit with ef-g(gdpnp) and rrf bound
|
| 76 | d1ldma1 |
|
not modelled |
64.3 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 77 | d1o6za1 |
|
not modelled |
64.1 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 78 | d1e0ta2 |
|
not modelled |
64.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 79 | d1nbca_ |
|
not modelled |
63.3 |
18 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
| 80 | d1a5za1 |
|
not modelled |
62.6 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 81 | d1llaa3 |
|
not modelled |
61.7 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Arthropod hemocyanin, C-terminal domain |
| 82 | c2b39B_ |
|
not modelled |
61.7 |
14 |
PDB header:immune system
Chain: B: PDB Molecule:c3;
PDBTitle: structure of mammalian c3 with an intact thioester at 3a resolution
|
| 83 | c1wziA_ |
|
not modelled |
60.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: structural basis for alteration of cofactor specificity of2 malate dehydrogenase from thermus flavus
|
| 84 | c1ogoX_ |
|
not modelled |
60.6 |
18 |
PDB header:hydrolase
Chain: X: PDB Molecule:dextranase;
PDBTitle: dex49a from penicillium minioluteum complex with isomaltose
|
| 85 | c2dfdD_ |
|
not modelled |
60.5 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of human malate dehydrogenase type 2
|
| 86 | c3cu7A_ |
|
not modelled |
60.1 |
13 |
PDB header:immune system
Chain: A: PDB Molecule:complement c5;
PDBTitle: human complement component 5
|
| 87 | d2ldxa1 |
|
not modelled |
59.5 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 88 | c2pwzG_ |
|
not modelled |
59.2 |
18 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of the apo form of e.coli malate dehydrogenase
|
| 89 | d1i0za1 |
|
not modelled |
58.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 90 | c2pn5A_ |
|
not modelled |
58.5 |
17 |
PDB header:immune system
Chain: A: PDB Molecule:thioester-containing protein i;
PDBTitle: crystal structure of tep1r
|
| 91 | c8ldhA_ |
|
not modelled |
58.5 |
19 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:m4 apo-lactate dehydrogenase;
PDBTitle: refined crystal structure of dogfish m4 apo-lactate2 dehydrogenase
|
| 92 | c1hyhA_ |
|
not modelled |
58.2 |
20 |
PDB header:oxidoreductase (choh(d)-nad+(a))
Chain: A: PDB Molecule:l-2-hydroxyisocaproate dehydrogenase;
PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
|
| 93 | d1jv2a2 |
|
not modelled |
56.9 |
22 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Integrin domains
Family:Integrin domains |
| 94 | d1p5va1 |
|
not modelled |
55.8 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:PapD-like
Family:Pilus chaperone |
| 95 | c2qv2A_ |
|
not modelled |
55.7 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:inositol polyphosphate 5-phosphatase ocrl-1;
PDBTitle: a role of the lowe syndrome protein ocrl in early steps of2 the endocytic pathway
|
| 96 | c1jz6C_ |
|
not modelled |
55.1 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-galactosidase;
PDBTitle: e. coli (lacz) beta-galactosidase in complex with galacto-2 tetrazole
|
| 97 | c3tl2A_ |
|
not modelled |
54.7 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of bacillus anthracis str. ames malate dehydrogenase2 in closed conformation.
|
| 98 | d5ldha1 |
|
not modelled |
54.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 99 | d1nwpa_ |
|
not modelled |
53.8 |
21 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 100 | c2hjrK_ |
|
not modelled |
53.2 |
18 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
| 101 | d1l4ia1 |
|
not modelled |
53.1 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:PapD-like
Family:Pilus chaperone |
| 102 | c2o14A_ |
|
not modelled |
52.6 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yxim;
PDBTitle: x-ray crystal structure of protein yxim_bacsu from bacillus2 subtilis. northeast structural genomics consortium target3 sr595
|
| 103 | c2l8aA_ |
|
not modelled |
52.6 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase;
PDBTitle: structure of a novel cbm3 lacking the calcium-binding site
|
| 104 | d1i10a1 |
|
not modelled |
52.5 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 105 | c2v6bB_ |
|
not modelled |
52.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of lactate dehydrogenase from deinococcus2 radiodurans (apo form)
|
| 106 | c2jtxA_ |
|
not modelled |
51.9 |
55 |
PDB header:transcription
Chain: A: PDB Molecule:transcription initiation factor iie subunit
PDBTitle: nmr structure of the tfiie-alpha carboxyl terminus
|
| 107 | c1lldA_ |
|
not modelled |
51.2 |
14 |
PDB header:oxidoreductase(choh (d)-nad (a))
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: molecular basis of allosteric activation of bacterial l-lactate2 dehydrogenase
|
| 108 | c3mpbA_ |
|
not modelled |
51.1 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar isomerase;
PDBTitle: z5688 from e. coli o157:h7 bound to fructose
|
| 109 | d1a3xa2 |
|
not modelled |
50.3 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 110 | d9ldta1 |
|
not modelled |
50.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 111 | c2rnqA_ |
|
not modelled |
49.5 |
55 |
PDB header:transcription
Chain: A: PDB Molecule:transcription initiation factor iie subunit
PDBTitle: solution structure of the c-terminal acidic domain of tfiie2 alpha
|
| 112 | c1a5zA_ |
|
not modelled |
49.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: lactate dehydrogenase from thermotoga maritima (tmldh)
|
| 113 | d1hyha1 |
|
not modelled |
48.8 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 114 | c1t5aB_ |
|
not modelled |
48.7 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase, m2 isozyme;
PDBTitle: human pyruvate kinase m2
|
| 115 | c3q48B_ |
|
not modelled |
48.6 |
13 |
PDB header:chaperone
Chain: B: PDB Molecule:chaperone cupb2;
PDBTitle: crystal structure of pseudomonas aeruginosa cupb2 chaperone
|
| 116 | c1z9sA_ |
|
not modelled |
47.9 |
16 |
PDB header:chaperone/immune system
Chain: A: PDB Molecule:chaperone protein caf1m;
PDBTitle: crystal structure of the native chaperone:subunit:subunit2 caf1m:caf1:caf1 complex
|
| 117 | d1iq8a1 |
|
not modelled |
47.5 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:tRNA-guanine transglycosylase
Family:tRNA-guanine transglycosylase |
| 118 | c3p7mC_ |
|
not modelled |
47.2 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of putative lactate dehydrogenase from francisella2 tularensis subsp. tularensis schu s4
|
| 119 | d1joia_ |
|
not modelled |
47.1 |
18 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 120 | c2e28A_ |
|
not modelled |
46.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
|