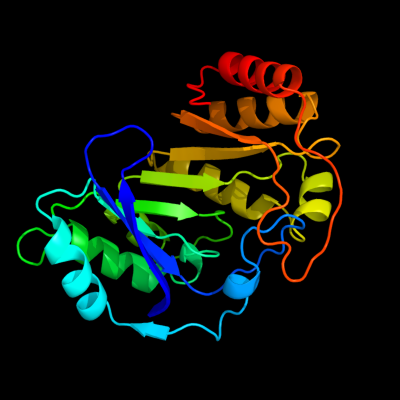
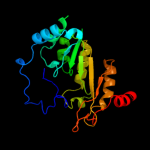
1 c3c8fA_
100.0
32
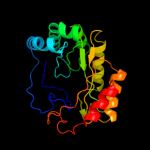
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate formate-lyase 1-activating enzyme;PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
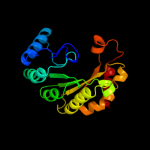
2 c3canA_
99.9
22
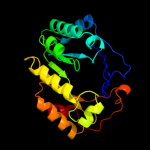
PDB header: lyase activatorChain: A: PDB Molecule: pyruvate-formate lyase-activating enzyme;PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
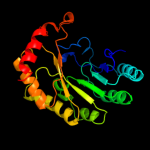
3 c3rfaA_
99.9
18
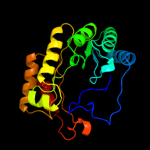
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
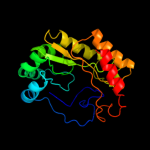
4 c2yx0A_
99.9
20
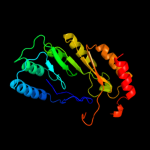
PDB header: metal binding proteinChain: A: PDB Molecule: radical sam enzyme;PDBTitle: crystal structure of p. horikoshii tyw1
5 d1tv8a_
99.7
20
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: MoCo biosynthesis proteins6 c2z2uA_
99.6
19
PDB header: metal binding proteinChain: A: PDB Molecule: upf0026 protein mj0257;PDBTitle: crystal structure of archaeal tyw1
7 c2a5hC_
99.4
15
PDB header: isomeraseChain: C: PDB Molecule: l-lysine 2,3-aminomutase;PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
8 d1r30a_
99.3
17
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Biotin synthase9 c1r30A_
99.3
17
PDB header: transferaseChain: A: PDB Molecule: biotin synthase;PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
10 c3t7vA_
99.2
11
PDB header: transferaseChain: A: PDB Molecule: methylornithine synthase pylb;PDBTitle: crystal structure of methylornithine synthase (pylb)
11 c3cixA_
99.0
19
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
12 d1olta_
98.9
10
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Oxygen-independent coproporphyrinogen III oxidase HemN13 d1bc6a_
98.6
23
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin14 d7fd1a_
98.6
19
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin15 c1gx7A_
98.5
22
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic [fe] hydrogenase large subunit;PDBTitle: best model of the electron transfer complex between2 cytochrome c3 and [fe]-hydrogenase
16 c2gmhA_
98.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: electron transfer flavoprotein-ubiquinonePDBTitle: structure of porcine electron transfer flavoprotein-2 ubiquinone oxidoreductase in complexed with ubiquinone
17 c1gthD_
98.5
28
PDB header: oxidoreductaseChain: D: PDB Molecule: dihydropyrimidine dehydrogenase;PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
18 c2v2kB_
98.5
16
PDB header: electron transportChain: B: PDB Molecule: ferredoxin;PDBTitle: the crystal structure of fdxa, a 7fe ferredoxin from2 mycobacterium smegmatis
19 c2v4jE_
98.5
20
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
20 c1hfeL_
98.5
21
PDB header: hydrogenaseChain: L: PDB Molecule: protein (fe-only hydrogenase (e.c.1.18.99.1)PDBTitle: 1.6 a resolution structure of the fe-only hydrogenase from2 desulfovibrio desulfuricans
21 c2zvsB_
not modelled
98.5
30
PDB header: electron transportChain: B: PDB Molecule: uncharacterized ferredoxin-like protein yfhl;PDBTitle: crystal structure of the 2[4fe-4s] ferredoxin from escherichia coli
22 d2fug91
not modelled
98.4
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins23 c2fugG_
not modelled
98.4
24
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase chain 9;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
24 d1h98a_
not modelled
98.4
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin25 c2ivfB_
not modelled
98.4
27
PDB header: oxidoreductaseChain: B: PDB Molecule: ethylbenzene dehydrogenase beta-subunit;PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
26 d1rgva_
not modelled
98.3
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins27 d1y5ib1
not modelled
98.3
27
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins28 c3c7bA_
not modelled
98.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunit alpha;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
29 d1blua_
not modelled
98.3
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins30 c3c7bE_
not modelled
98.3
21
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunit beta;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
31 d1fcaa_
not modelled
98.3
33
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins32 c3gyxJ_
not modelled
98.3
29
PDB header: oxidoreductaseChain: J: PDB Molecule: adenylylsulfate reductase;PDBTitle: crystal structure of adenylylsulfate reductase from2 desulfovibrio gigas
33 c2fgoA_
not modelled
98.2
21
PDB header: electron transportChain: A: PDB Molecule: ferredoxin;PDBTitle: structure of the 2[4fe-4s] ferredoxin from pseudomonas2 aeruginosa
34 d2fdna_
not modelled
98.2
35
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins35 c1ti2F_
not modelled
98.2
22
PDB header: oxidoreductaseChain: F: PDB Molecule: pyrogallol hydroxytransferase small subunit;PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici
36 d1jnrb_
not modelled
98.2
27
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins37 d1hfel2
not modelled
98.2
33
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins38 c2vpyB_
not modelled
98.2
17
PDB header: oxidoreductaseChain: B: PDB Molecule: nrfc protein;PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
39 d1clfa_
not modelled
98.2
32
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins40 c2v4jA_
not modelled
98.2
25
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
41 d1jb0c_
not modelled
98.1
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin42 c1c4cA_
not modelled
98.1
21
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (fe-only hydrogenase);PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
43 c2c3yA_
not modelled
98.1
21
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate-ferredoxin oxidoreductase;PDBTitle: crystal structure of the radical form of2 pyruvate:ferredoxin oxidoreductase from desulfovibrio3 africanus
44 d1kqfb1
not modelled
98.1
20
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins45 d1h0hb_
not modelled
98.0
14
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins46 d1dura_
not modelled
98.0
30
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins47 d1xera_
not modelled
98.0
27
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Archaeal ferredoxins48 c1kqfB_
not modelled
98.0
26
PDB header: oxidoreductaseChain: B: PDB Molecule: formate dehydrogenase, nitrate-inducible, iron-sulfurPDBTitle: formate dehydrogenase n from e. coli
49 d1gtea5
not modelled
98.0
26
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins50 c2qgqF_
not modelled
97.9
13
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: protein tm_1862;PDBTitle: crystal structure of tm_1862 from thermotoga maritima.2 northeast structural genomics consortium target vr77
51 d2gmha3
not modelled
97.9
25
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: ETF-QO domain-like52 d3c7bb1
not modelled
97.6
20
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins53 d2c42a5
not modelled
97.5
20
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins54 d3c8ya3
not modelled
97.5
23
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins55 d2fug34
not modelled
97.4
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins56 d1vlfn2
not modelled
97.4
22
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins57 d1vjwa_
not modelled
97.4
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin58 d1iqza_
not modelled
97.4
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin59 c2fugC_
not modelled
96.9
17
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh-quinone oxidoreductase chain 3;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
60 d1sj1a_
not modelled
96.9
21
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin61 c3bk7A_
not modelled
96.6
29
PDB header: hydrolyase/translationChain: A: PDB Molecule: abc transporter atp-binding protein;PDBTitle: structure of the complete abce1/rnaase-l inhibitor protein2 from pyrococcus abysii
62 c1dwlA_
not modelled
96.0
27
PDB header: electron transferChain: A: PDB Molecule: ferredoxin i;PDBTitle: the ferredoxin-cytochrome complex using heteronuclear nmr2 and docking simulation
63 d1fxra_
not modelled
95.9
17
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin64 d1kf6b1
not modelled
94.4
28
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain65 d2bs2b1
not modelled
94.4
22
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain66 c3cf4A_
not modelled
94.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: acetyl-coa decarboxylase/synthase alpha subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex
67 c1nekB_
not modelled
93.7
20
PDB header: oxidoreductase/electron transportChain: B: PDB Molecule: succinate dehydrogenase iron-sulfur protein;PDBTitle: complex ii (succinate dehydrogenase) from e. coli with2 ubiquinone bound
68 c2bs2E_
not modelled
93.5
23
PDB header: oxidoreductaseChain: E: PDB Molecule: quinol-fumarate reductase iron-sulfur subunit b;PDBTitle: quinol:fumarate reductase from wolinella succinogenes
69 d1nekb1
not modelled
93.3
23
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain70 c2b76N_
not modelled
93.0
29
PDB header: oxidoreductaseChain: N: PDB Molecule: fumarate reductase iron-sulfur protein;PDBTitle: e. coli quinol fumarate reductase frda e49q mutation
71 d2v4ja1
not modelled
92.6
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins72 c2h89B_
not modelled
92.4
20
PDB header: oxidoreductaseChain: B: PDB Molecule: succinate dehydrogenase ip subunit;PDBTitle: avian respiratory complex ii with malonate bound
73 d3c7ba1
not modelled
92.0
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins74 c1gcyA_
not modelled
90.9
17
PDB header: hydrolaseChain: A: PDB Molecule: glucan 1,4-alpha-maltotetrahydrolase;PDBTitle: high resolution crystal structure of maltotetraose-forming2 exo-amylase
75 c1jgiA_
not modelled
90.8
17
PDB header: transferaseChain: A: PDB Molecule: amylosucrase;PDBTitle: crystal structure of the active site mutant glu328gln of2 amylosucrase from neisseria polysaccharea in complex with3 the natural substrate sucrose
76 d1m53a2
not modelled
90.1
21
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain77 d1g5aa2
not modelled
89.5
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain78 d2v4jb1
not modelled
89.4
24
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins79 c3a47A_
not modelled
89.2
19
PDB header: hydrolaseChain: A: PDB Molecule: oligo-1,6-glucosidase;PDBTitle: crystal structure of isomaltase from saccharomyces cerevisiae
80 c1m7xC_
not modelled
88.8
21
PDB header: transferaseChain: C: PDB Molecule: 1,4-alpha-glucan branching enzyme;PDBTitle: the x-ray crystallographic structure of branching enzyme
81 c1m53A_
not modelled
88.1
21
PDB header: isomeraseChain: A: PDB Molecule: isomaltulose synthase;PDBTitle: crystal structure of isomaltulose synthase (pali) from2 klebsiella sp. lx3
82 c1ehaA_
not modelled
87.5
15
PDB header: hydrolaseChain: A: PDB Molecule: glycosyltrehalose trehalohydrolase;PDBTitle: crystal structure of glycosyltrehalose trehalohydrolase2 from sulfolobus solfataricus
83 c3k1dA_
not modelled
87.2
31
PDB header: transferaseChain: A: PDB Molecule: 1,4-alpha-glucan-branching enzyme;PDBTitle: crystal structure of glycogen branching enzyme synonym: 1,4-alpha-d-2 glucan:1,4-alpha-d-glucan 6-glucosyl-transferase from mycobacterium3 tuberculosis h37rv
84 c3czkA_
not modelled
86.8
21
PDB header: hydrolaseChain: A: PDB Molecule: sucrose hydrolase;PDBTitle: crystal structure analysis of sucrose hydrolase(suh) e322q-2 sucrose complex
85 d1uoka2
not modelled
86.8
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain86 c1cygA_
not modelled
86.8
20
PDB header: glycosyltransferaseChain: A: PDB Molecule: cyclodextrin glucanotransferase;PDBTitle: cyclodextrin glucanotransferase (e.c.2.4.1.19) (cgtase)
87 c1bagA_
not modelled
86.8
7
PDB header: alpha-amylaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: alpha-amylase from bacillus subtilis complexed with2 maltopentaose
88 c3k8kB_
not modelled
86.5
19
PDB header: membrane proteinChain: B: PDB Molecule: alpha-amylase, susg;PDBTitle: crystal structure of susg
89 c3blpX_
not modelled
86.4
12
PDB header: hydrolaseChain: X: PDB Molecule: alpha-amylase 1;PDBTitle: role of aromatic residues in human salivary alpha-amylase
90 c1zjaB_
not modelled
86.3
25
PDB header: isomeraseChain: B: PDB Molecule: trehalulose synthase;PDBTitle: crystal structure of the trehalulose synthase mutb from2 pseudomonas mesoacidophila mx-45 (triclinic form)
91 c1tcmB_
not modelled
86.2
17
PDB header: glycosyltransferaseChain: B: PDB Molecule: cyclodextrin glycosyltransferase;PDBTitle: cyclodextrin glycosyltransferase w616a mutant from bacillus2 circulans strain 251
92 c1qhoA_
not modelled
86.0
14
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: five-domain alpha-amylase from bacillus stearothermophilus,2 maltose/acarbose complex
93 c2ze0A_
not modelled
85.6
20
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: alpha-glucosidase gsj
94 d1gcya2
not modelled
85.5
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain95 c1uokA_
not modelled
85.3
19
PDB header: glucosidaseChain: A: PDB Molecule: oligo-1,6-glucosidase;PDBTitle: crystal structure of b. cereus oligo-1,6-glucosidase
96 c3amkA_
not modelled
85.2
24
PDB header: transferaseChain: A: PDB Molecule: os06g0726400 protein;PDBTitle: structure of the starch branching enzyme i (bei) from oryza sativa l
97 d1ua7a2
not modelled
84.9
7
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain98 c3m07A_
not modelled
84.5
21
PDB header: unknown functionChain: A: PDB Molecule: putative alpha amylase;PDBTitle: 1.4 angstrom resolution crystal structure of putative alpha2 amylase from salmonella typhimurium.
99 c3bmwA_
not modelled
84.4
13
PDB header: transferaseChain: A: PDB Molecule: cyclomaltodextrin glucanotransferase;PDBTitle: cyclodextrin glycosyl transferase from thermoanerobacterium2 thermosulfurigenes em1 mutant s77p complexed with a maltoheptaose3 inhibitor
100 c2by0A_
not modelled
84.4
28
PDB header: hydrolaseChain: A: PDB Molecule: maltooligosyltrehalose trehalohydrolase;PDBTitle: is radiation damage dependent on the dose-rate used during2 macromolecular crystallography data collection
101 d1gjwa2
not modelled
84.4
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain102 c3amlA_
not modelled
84.3
26
PDB header: transferaseChain: A: PDB Molecule: os06g0726400 protein;PDBTitle: structure of the starch branching enzyme i (bei) from oryza sativa l
103 d2bhua3
not modelled
83.8
21
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain104 d1m7xa3
not modelled
83.6
21
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain105 c2dh3A_
not modelled
82.7
13
PDB header: transport protein, signaling proteinChain: A: PDB Molecule: 4f2 cell-surface antigen heavy chain;PDBTitle: crystal structure of human ed-4f2hc
106 d1gvia3
not modelled
82.1
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain107 c2ya0A_
not modelled
82.0
15
PDB header: hydrolaseChain: A: PDB Molecule: putative alkaline amylopullulanase;PDBTitle: catalytic module of the multi-modular glycogen-degrading2 pneumococcal virulence factor spua
108 d1wzla3
not modelled
81.9
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain109 d1ea9c3
not modelled
81.9
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain110 c1jaeA_
not modelled
81.6
14
PDB header: glycosidaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: structure of tenebrio molitor larval alpha-amylase
111 c1jd7A_
not modelled
81.0
7
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: crystal structure analysis of the mutant k300r of2 pseudoalteromonas haloplanctis alpha-amylase
112 d1j0ha3
not modelled
80.9
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain113 c2wskA_
not modelled
80.8
16
PDB header: hydrolaseChain: A: PDB Molecule: glycogen debranching enzyme;PDBTitle: crystal structure of glycogen debranching enzyme glgx from2 escherichia coli k-12
114 c1jdaA_
not modelled
80.6
17
PDB header: hydrolaseChain: A: PDB Molecule: 1,4-alpha maltotetrahydrolase;PDBTitle: maltotetraose-forming exo-amylase
115 c3zt5D_
not modelled
80.6
16
PDB header: hydrolaseChain: D: PDB Molecule: putative glucanohydrolase pep1a;PDBTitle: glge isoform 1 from streptomyces coelicolor with maltose2 bound
116 c1jibA_
not modelled
80.1
14
PDB header: hydrolaseChain: A: PDB Molecule: neopullulanase;PDBTitle: complex of alpha-amylase ii (tva ii) from thermoactinomyces2 vulgaris r-47 with maltotetraose based on a crystal soaked3 with maltohexaose.
117 d1h3ga3
not modelled
79.7
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain118 c2zidA_
not modelled
79.7
21
PDB header: hydrolaseChain: A: PDB Molecule: dextran glucosidase;PDBTitle: crystal structure of dextran glucosidase e236q complex with2 isomaltotriose
119 c3edeB_
not modelled
79.2
20
PDB header: hydrolaseChain: B: PDB Molecule: cyclomaltodextrinase;PDBTitle: structural base for cyclodextrin hydrolysis
120 c3bzjA_
not modelled
78.8
18
PDB header: hydrolaseChain: A: PDB Molecule: uv endonuclease;PDBTitle: uvde k229l