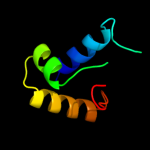
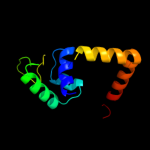
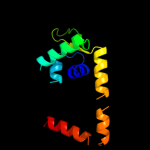
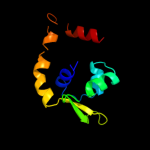
1 c3tpaA_
100.0
17
PDB header: heme binding proteinChain: A: PDB Molecule: heme-binding protein a;PDBTitle: structure of hbpa2 from haemophilus parasuis
2 d1dpea_
100.0
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like3 c2wokA_
100.0
18
PDB header: peptide binding protein/peptideChain: A: PDB Molecule: clavulanic acid biosynthesis oligopeptidePDBTitle: clavulanic acid biosynthesis oligopeptide2 binding protein 2 complexed with bradykinin
4 d1jeta_
100.0
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like5 c3m8uA_
100.0
16
PDB header: transport proteinChain: A: PDB Molecule: heme-binding protein a;PDBTitle: crystal structure of glutathione-binding protein a (gbpa) from2 haemophilus parasuis sh0165 in complex with glutathione disulfide3 (gssg)
6 d1uqwa_
100.0
19
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like7 c3o9pA_
100.0
16
PDB header: peptide binding protein/peptideChain: A: PDB Molecule: periplasmic murein peptide-binding protein;PDBTitle: the structure of the escherichia coli murein tripeptide binding2 protein mppa
8 d1zlqa1
100.0
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like9 d1xoca1
100.0
19
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like10 c3t66A_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: nickel abc transporter (nickel-binding protein);PDBTitle: crystal structure of nickel abc transporter from bacillus halodurans
11 c3rqtA_
100.0
13
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: 1.5 angstrom crystal structure of the complex of ligand binding2 component of abc-type import system from staphylococcus aureus with3 nickel and two histidines
12 c3ry3B_
100.0
16
PDB header: transport proteinChain: B: PDB Molecule: putative solute-binding protein;PDBTitle: putative solute-binding protein from yersinia pestis.
13 d1vr5a1
100.0
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like14 c2o7jA_
100.0
13
PDB header: sugar binding proteinChain: A: PDB Molecule: oligopeptide abc transporter, periplasmicPDBTitle: the x-ray crystal structure of a thermophilic cellobiose2 binding protein bound with cellopentaose
15 c2grvC_
100.0
13
PDB header: biosynthetic proteinChain: C: PDB Molecule: lpqw;PDBTitle: crystal structure of lpqw
16 c3ftoA_
100.0
13
PDB header: peptide binding proteinChain: A: PDB Molecule: oligopeptide-binding protein oppa;PDBTitle: crystal structure of oppa in a open conformation
17 c1ztyA_
100.0
15
PDB header: sugar binding protein, signaling proteinChain: A: PDB Molecule: chitin oligosaccharide binding protein;PDBTitle: crystal structure of the chitin oligasaccharide binding2 protein
18 c2d5wA_
100.0
13
PDB header: peptide binding proteinChain: A: PDB Molecule: peptide abc transporter, peptide-binding protein;PDBTitle: the crystal structure of oligopeptide binding protein from thermus2 thermophilus hb8 complexed with pentapeptide
19 c3pamB_
99.8
16
PDB header: transport proteinChain: B: PDB Molecule: transmembrane protein;PDBTitle: crystal structure of a domain of transmembrane protein of abc-type2 oligopeptide transport system from bartonella henselae str. houston-1
20 c3lvuB_
99.8
15
PDB header: transport proteinChain: B: PDB Molecule: abc transporter, periplasmic substrate-binding protein;PDBTitle: crystal structure of abc transporter, periplasmic substrate-binding2 protein spo2066 from silicibacter pomeroyi
21 c3o6pA_
not modelled
99.7
16
PDB header: protein bindingChain: A: PDB Molecule: peptide abc transporter, peptide-binding protein;PDBTitle: crystal structure of peptide abc transporter, peptide-binding protein
22 d1lnwa_
97.3
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators23 d1i5za1
97.1
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like24 d1sfxa_
97.1
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: TrmB-like25 d2oz6a1
97.1
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like26 c3g3zA_
97.0
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
27 c3bj6B_
97.0
13
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of marr family transcription regulator sp03579
28 d2h6ca1
not modelled
96.9
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like29 d2gaua1
not modelled
96.8
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like30 d1biaa1
not modelled
96.8
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like31 c3nqoB_
not modelled
96.8
11
PDB header: transcriptionChain: B: PDB Molecule: marr-family transcriptional regulator;PDBTitle: crystal structure of a marr family transcriptional regulator (cd1569)2 from clostridium difficile 630 at 2.20 a resolution
32 d1hsja1
not modelled
96.8
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators33 c2gxgA_
not modelled
96.7
10
PDB header: transcriptionChain: A: PDB Molecule: 146aa long hypothetical transcriptional regulator;PDBTitle: crystal structure of emrr homolog from hyperthermophilic archaea2 sulfolobus tokodaii strain7
34 d2fbha1
not modelled
96.7
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators35 d2etha1
not modelled
96.7
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators36 d1lj9a_
not modelled
96.7
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators37 d3broa1
96.7
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators38 d1s3ja_
not modelled
96.6
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators39 c3fm5D_
not modelled
96.6
13
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator;PDBTitle: x-ray crystal structure of transcriptional regulator (marr family)2 from rhodococcus sp. rha1
40 d1ft9a1
not modelled
96.6
32
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like41 d1p4xa1
not modelled
96.6
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators42 c1ft9A_
not modelled
96.6
28
PDB header: transcriptionChain: A: PDB Molecule: carbon monoxide oxidation system transcriptionPDBTitle: structure of the reduced (feii) co-sensing protein from r.2 rubrum
43 c3r0aB_
not modelled
96.6
5
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator;PDBTitle: possible transcriptional regulator from methanosarcina mazei go1 (gi2 21227196)
44 c3dv8A_
not modelled
96.5
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, crp/fnr family;PDBTitle: crystal structure of a putative transcriptional regulator of the2 crp/fnr family (eubrec_1222) from eubacterium rectale atcc 33656 at3 2.55 a resolution
45 c3ctaA_
not modelled
96.5
18
PDB header: transferaseChain: A: PDB Molecule: riboflavin kinase;PDBTitle: crystal structure of riboflavin kinase from thermoplasma2 acidophilum
46 d2coha1
not modelled
96.5
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like47 c3bjaA_
not modelled
96.4
12
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, marr family, putative;PDBTitle: crystal structure of putative marr-like transcription regulator2 (np_978771.1) from bacillus cereus atcc 10987 at 2.38 a resolution
48 c2v79B_
not modelled
96.4
19
PDB header: dna-binding proteinChain: B: PDB Molecule: dna replication protein dnad;PDBTitle: crystal structure of the n-terminal domain of dnad from2 bacillus subtilis
49 c1fx7C_
not modelled
96.3
15
PDB header: signaling proteinChain: C: PDB Molecule: iron-dependent repressor ider;PDBTitle: crystal structure of the iron-dependent regulator (ider)2 from mycobacterium tuberculosis
50 c3hrmA_
not modelled
96.3
12
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional regulator sarz;PDBTitle: crystal structure of staphylococcus aureus protein sarz in sulfenic2 acid form
51 d2frha1
not modelled
96.3
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators52 c3f3xA_
not modelled
96.3
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, marr family, putative;PDBTitle: crystal structure of the transcriptional regulator bldr2 from sulfolobus solfataricus
53 d1p4xa2
not modelled
96.2
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators54 d1j5ya1
not modelled
96.2
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like55 c3e6mD_
not modelled
96.2
14
PDB header: transcription regulatorChain: D: PDB Molecule: marr family transcriptional regulator;PDBTitle: the crystal structure of a marr family transcriptional2 regulator from silicibacter pomeroyi dss.
56 d2a61a1
not modelled
96.2
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators57 d3ctaa1
not modelled
96.1
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators58 c2oz6A_
not modelled
96.0
18
PDB header: dna binding proteinChain: A: PDB Molecule: virulence factor regulator;PDBTitle: crystal structure of virulence factor regulator from pseudomonas2 aeruginosa in complex with camp
59 c2nnnB_
not modelled
96.0
18
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulator;PDBTitle: crystal structure of probable transcriptional regulator from2 pseudomonas aeruginosa
60 c3cjnA_
not modelled
96.0
14
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of transcriptional regulator, marr family, from2 silicibacter pomeroyi
61 c2gauA_
not modelled
95.9
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, crp/fnr family;PDBTitle: crystal structure of transcriptional regulator, crp/fnr family from2 porphyromonas gingivalis (apc80792), structural genomics, mcsg
62 c3k0lA_
not modelled
95.9
18
PDB header: transcription regulatorChain: A: PDB Molecule: repressor protein;PDBTitle: crystal structure of putative marr family transcriptional2 regulator from acinetobacter sp. adp
63 d1jgsa_
not modelled
95.8
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators64 c3iwzB_
not modelled
95.8
27
PDB header: transcriptionChain: B: PDB Molecule: catabolite activation-like protein;PDBTitle: the c-di-gmp responsive global regulator clp links cell-cell signaling2 to virulence gene expression in xanthomonas campestris
65 c1p4xA_
not modelled
95.8
6
PDB header: transcriptionChain: A: PDB Molecule: staphylococcal accessory regulator a homologue;PDBTitle: crystal structure of sars protein from staphylococcus aureus
66 d1jhfa1
not modelled
95.7
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain67 d3e5ua1
not modelled
95.7
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like68 d2fbia1
not modelled
95.7
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators69 c3neuA_
not modelled
95.7
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin1836 protein;PDBTitle: the crystal structure of a functionally-unknown protein lin1836 from2 listeria innocua clip11262
70 c3e6dA_
not modelled
95.7
5
PDB header: transcription regulationChain: A: PDB Molecule: cyclic nucleotide-binding protein;PDBTitle: crystal structure of cprk c200s
71 c2x4hA_
not modelled
95.6
11
PDB header: transcriptionChain: A: PDB Molecule: hypothetical protein sso2273;PDBTitle: crystal structure of the hypothetical protein sso2273 from2 sulfolobus solfataricus
72 c2rdpA_
not modelled
95.5
13
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator marr;PDBTitle: the structure of a marr family protein from bacillus2 stearothermophilus
73 d2bgca1
not modelled
95.5
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like74 c3by6C_
not modelled
95.4
11
PDB header: transcription regulatorChain: C: PDB Molecule: predicted transcriptional regulator;PDBTitle: crystal structure of a transcriptional regulator from oenococcus oeni
75 c2fxaB_
not modelled
95.4
12
PDB header: transcriptionChain: B: PDB Molecule: protease production regulatory protein hpr;PDBTitle: structure of the protease production regulatory protein hpr from2 bacillus subtilis.
76 c1zybA_
not modelled
95.4
26
PDB header: transcription regulatorChain: A: PDB Molecule: transcription regulator, crp family;PDBTitle: crystal structure of transcription regulator from bacteroides2 thetaiotaomicron vpi-5482 at 2.15 a resolution
77 d1ub9a_
not modelled
95.4
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators78 c2zdbA_
not modelled
95.4
13
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, crp family;PDBTitle: crystal structure of tthb099, a transcriptional regulator crp family2 from thermus thermophilus hb8
79 c3bpxB_
not modelled
95.3
13
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of marr
80 c1g3wA_
not modelled
95.3
11
PDB header: gene regulationChain: A: PDB Molecule: diphtheria toxin repressor;PDBTitle: cd-cys102ser dtxr
81 c3d0sA_
not modelled
95.2
29
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: camp receptor protein from m.tuberculosis, camp-free form
82 c2it0A_
not modelled
95.2
12
PDB header: transcription/dnaChain: A: PDB Molecule: iron-dependent repressor ider;PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
83 c2fa5B_
not modelled
95.2
8
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator marr/emrr family;PDBTitle: the crystal structure of an unliganded multiple antibiotic-2 resistance repressor (marr) from xanthomonas campestris
84 d2isya1
not modelled
95.2
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein85 d2bv6a1
not modelled
95.2
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators86 c2nyxB_
not modelled
95.2
13
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulatory protein, rv1404;PDBTitle: crystal structure of rv1404 from mycobacterium tuberculosis
87 c3kccA_
not modelled
95.0
22
PDB header: transcriptionChain: A: PDB Molecule: catabolite gene activator;PDBTitle: crystal structure of d138l mutant of catabolite gene activator protein
88 c1f5tA_
not modelled
95.0
11
PDB header: transcription/dnaChain: A: PDB Molecule: diphtheria toxin repressor;PDBTitle: diphtheria tox repressor (c102d mutant) complexed with2 nickel and dtxr consensus binding sequence
89 d2hr3a1
not modelled
95.0
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators90 c2zcwA_
not modelled
94.9
18
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, fnr/crp family;PDBTitle: crystal structure of ttha1359, a transcriptional regulator,2 crp/fnr family from thermus thermophilus hb8
91 c3hruA_
not modelled
94.9
15
PDB header: transcriptionChain: A: PDB Molecule: metalloregulator scar;PDBTitle: crystal structure of scar with bound zn2+
92 d1mkma1
not modelled
94.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator IclR, N-terminal domain93 d1g3wa1
not modelled
94.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein94 c2y75F_
not modelled
94.7
15
PDB header: transcriptionChain: F: PDB Molecule: hth-type transcriptional regulator cymr;PDBTitle: the structure of cymr (yrzc) the global cysteine regulator2 of b. subtilis
95 c3e97A_
not modelled
94.6
11
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, crp/fnr family;PDBTitle: crystal structure of transcriptional regulator of crp/fnr2 family (yp_604437.1) from deinococcus geothermalis dsm3 11300 at 1.86 a resolution
96 d1stza1
not modelled
94.6
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain97 c3s2wB_
not modelled
94.5
17
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the crystal structure of a marr transcriptional regulator from2 methanosarcina mazei go1
98 d2fbka1
not modelled
94.5
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators99 c3dkwB_
not modelled
94.4
15
PDB header: transcription regulatorChain: B: PDB Molecule: dnr protein;PDBTitle: crystal structure of dnr from pseudomonas aeruginosa.
100 d1zyba1
not modelled
94.4
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like101 d1u2wa1
not modelled
94.4
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators102 c2bgcA_
not modelled
94.4
13
PDB header: transcriptionChain: A: PDB Molecule: prfa;PDBTitle: prfa-g145s, a constitutive active mutant of the2 transcriptional regulator in l.monocytogenes
103 c2vn2B_
not modelled
94.3
16
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
104 d1ku9a_
not modelled
94.2
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DNA-binding protein Mj223105 c2qwwB_
not modelled
94.2
8
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
106 c3deuB_
not modelled
94.2
16
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator slya;PDBTitle: crystal structure of transcription regulatory protein slya2 from salmonella typhimurium in complex with salicylate3 ligands
107 c1zreB_
not modelled
94.2
18
PDB header: gene regulation/dnaChain: B: PDB Molecule: catabolite gene activator;PDBTitle: 4 crystal structures of cap-dna with all base-pair2 substitutions at position 6, cap-[6g;17c]icap38 dna
108 d2ev0a1
not modelled
94.1
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein109 c3oopA_
not modelled
94.1
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin2960 protein;PDBTitle: the structure of a protein with unknown function from listeria innocua2 clip11262
110 c2fmyB_
not modelled
94.1
23
PDB header: dna binding proteinChain: B: PDB Molecule: carbon monoxide oxidation system transcription regulatorPDBTitle: co-dependent transcription factor cooa from carboxydothermus2 hydrogenoformans (imidazole-bound form)
111 c2o0yB_
not modelled
94.1
22
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator rha1_ro069532 (iclr-family) from rhodococcus sp.
112 c3nrvC_
not modelled
94.0
6
PDB header: transcription regulatorChain: C: PDB Molecule: putative transcriptional regulator (marr/emrr family);PDBTitle: crystal structure of marr/emrr family transcriptional regulator from2 acinetobacter sp. adp1
113 d1z91a1
not modelled
94.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators114 d1z05a1
not modelled
93.9
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ROK associated domain115 c3la2A_
not modelled
93.9
13
PDB header: transcriptionChain: A: PDB Molecule: global nitrogen regulator;PDBTitle: crystal structure of ntca in complex with 2-oxoglutarate
116 c2h09A_
not modelled
93.8
16
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator mntr;PDBTitle: crystal structure of diphtheria toxin repressor like protein2 from e. coli
117 c3kp3B_
not modelled
93.8
9
PDB header: transcription regulator/antibioticChain: B: PDB Molecule: transcriptional regulator tcar;PDBTitle: staphylococcus epidermidis in complex with ampicillin
118 c1mkmA_
not modelled
93.5
12
PDB header: transcriptionChain: A: PDB Molecule: iclr transcriptional regulator;PDBTitle: crystal structure of the thermotoga maritima iclr
119 d3deua1
not modelled
93.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators120 d3bwga1
not modelled
93.5
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: GntR-like transcriptional regulators