1 c3oaaC_
100.0
100
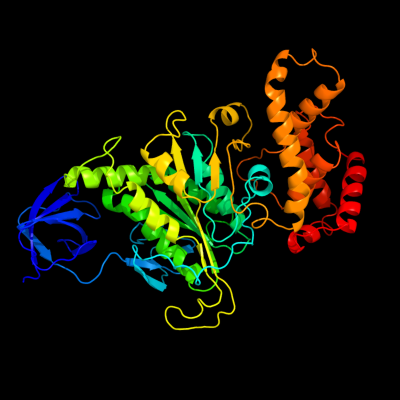
PDB header: hydrolase/transport proteinChain: C: PDB Molecule: atp synthase subunit alpha;PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
2 c2r9vA_
100.0
56
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase subunit alpha;PDBTitle: crystal structure of atp synthase subunit alpha (tm1612) from2 thermotoga maritima at 2.10 a resolution
3 c2w6fA_
100.0
60
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase subunit alpha heart isoform,PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 2.
4 c1kmhA_
100.0
55
PDB header: hydrolaseChain: A: PDB Molecule: atpase alpha subunit;PDBTitle: crystal structure of spinach chloroplast f1-atpase2 complexed with tentoxin
5 c1w0jB_
100.0
60
PDB header: hydrolaseChain: B: PDB Molecule: atp synthase alpha chain heart isoform,PDBTitle: beryllium fluoride inhibited bovine f1-atpase
6 c2qe7C_
100.0
59
PDB header: hydrolaseChain: C: PDB Molecule: atp synthase subunit alpha;PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
7 c3a5dB_
100.0
21
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase alpha chain;PDBTitle: inter-subunit interaction and quaternary rearrangement2 defined by the central stalk of prokaryotic v1-atpase
8 c3a5dM_
100.0
23
PDB header: hydrolaseChain: M: PDB Molecule: v-type atp synthase beta chain;PDBTitle: inter-subunit interaction and quaternary rearrangement2 defined by the central stalk of prokaryotic v1-atpase
9 c1fx0B_
100.0
25
PDB header: hydrolaseChain: B: PDB Molecule: atp synthase beta chain;PDBTitle: crystal structure of the chloroplast f1-atpase from spinach
10 c2dpyA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: flagellum-specific atp synthase;PDBTitle: crystal structure of the flagellar type iii atpase flii
11 c2w6jD_
100.0
24
PDB header: hydrolaseChain: D: PDB Molecule: atp synthase subunit beta, mitochondrial;PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 5.
12 c2jizD_
100.0
24
PDB header: hydrolaseChain: D: PDB Molecule: atp synthase subunit beta;PDBTitle: the structure of f1-atpase inhibited by resveratrol.
13 c1skyE_
100.0
26
PDB header: atp synthaseChain: E: PDB Molecule: f1-atpase;PDBTitle: crystal structure of the nucleotide free alpha3beta3 sub-complex of2 f1-atpase from the thermophilic bacillus ps3
14 c2c61A_
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: a-type atp synthase non-catalytic subunit b;PDBTitle: crystal structure of the non-catalytic b subunit of a-type2 atpase from m. mazei go1
15 c2oblA_
100.0
27
PDB header: hydrolaseChain: A: PDB Molecule: escn;PDBTitle: structural and biochemical analysis of a prototypical atpase from the2 type iii secretion system of pathogenic bacteria
16 c1vdzA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: a-type atpase subunit a;PDBTitle: crystal structure of a-type atpase catalytic subunit a from2 pyrococcus horikoshii ot3
17 d2jdia3
100.0
72
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)18 d1skyb3
100.0
68
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)19 d1fx0a3
100.0
66
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)20 d2jdid3
100.0
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)21 d1skye3
not modelled
100.0
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)22 d1fx0b3
not modelled
100.0
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)23 c3l0oB_
not modelled
100.0
18
PDB header: hydrolaseChain: B: PDB Molecule: transcription termination factor rho;PDBTitle: structure of rna-free rho transcription termination factor from2 thermotoga maritima
24 c1xpuB_
not modelled
100.0
16
PDB header: transcription/rnaChain: B: PDB Molecule: rho transcription termination factor;PDBTitle: structural mechanism of inhibition of the rho transcription2 termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-3 dihydrobicyclomycin (fpdb)
25 d1xpua3
not modelled
100.0
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)26 d1fx0a1
not modelled
99.9
38
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase27 d2jdia1
not modelled
99.9
47
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase28 d1skyb1
not modelled
99.9
35
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase29 d2f43a1
not modelled
99.9
47
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase30 d1skyb2
not modelled
99.5
40
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase31 d1maba2
99.5
35
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase32 d2jdia2
not modelled
99.5
38
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase33 d1fx0a2
not modelled
99.4
46
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase34 d1skye1
not modelled
98.7
13
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase35 c1u9iA_
not modelled
98.7
14
PDB header: circadian clock proteinChain: A: PDB Molecule: kaic;PDBTitle: crystal structure of circadian clock protein kaic with phosphorylation2 sites
36 d1fx0b1
not modelled
98.7
14
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase37 d2jdid1
not modelled
98.6
15
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase38 d1tf7a2
not modelled
98.3
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)39 c2recB_
not modelled
98.3
18
PDB header: helicasePDB COMPND: 40 d1tf7a1
not modelled
98.3
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)41 d1xp8a1
not modelled
98.2
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)42 c2zroA_
not modelled
98.2
18
PDB header: hydrolaseChain: A: PDB Molecule: protein reca;PDBTitle: msreca adp form iv
43 c3hr8A_
not modelled
98.2
19
PDB header: recombinationChain: A: PDB Molecule: protein reca;PDBTitle: crystal structure of thermotoga maritima reca
44 d1mo6a1
not modelled
98.1
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)45 c1xp8A_
not modelled
98.1
21
PDB header: dna binding proteinChain: A: PDB Molecule: reca protein;PDBTitle: "deinococcus radiodurans reca in complex with atp-gamma-s"
46 d1ubea1
not modelled
98.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)47 c3ldaA_
not modelled
98.0
19
PDB header: dna binding proteinChain: A: PDB Molecule: dna repair protein rad51;PDBTitle: yeast rad51 h352y filament interface mutant
48 c2dr3A_
not modelled
98.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0273 protein ph0284;PDBTitle: crystal structure of reca superfamily atpase ph0284 from pyrococcus2 horikoshii ot3
49 d2i1qa2
not modelled
98.0
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)50 c2w0mA_
not modelled
98.0
21
PDB header: unknown functionChain: A: PDB Molecule: sso2452;PDBTitle: crystal structure of sso2452 from sulfolobus solfataricus2 p2
51 d1n0wa_
not modelled
97.9
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)52 d1u94a1
not modelled
97.8
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)53 c2ztsB_
not modelled
97.8
15
PDB header: atp-binding proteinChain: B: PDB Molecule: putative uncharacterized protein ph0186;PDBTitle: crystal structure of kaic-like protein ph0186 from2 hyperthermophilic archaea pyrococcus horikoshii ot3
54 c2dflA_
not modelled
97.8
18
PDB header: recombinationChain: A: PDB Molecule: dna repair and recombination protein rada;PDBTitle: crystal structure of left-handed rada filament
55 c1t4gA_
not modelled
97.8
18
PDB header: recombinationChain: A: PDB Molecule: dna repair and recombination protein rada;PDBTitle: atpase in complex with amp-pnp
56 d1v5wa_
not modelled
97.8
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)57 d1pzna2
not modelled
97.7
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)58 c3io5B_
not modelled
97.7
14
PDB header: dna binding proteinChain: B: PDB Molecule: recombination and repair protein;PDBTitle: crystal structure of a dimeric form of the uvsx recombinase core2 domain from enterobacteria phage t4
59 d1szpa2
not modelled
97.7
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)60 c1szpC_
not modelled
97.6
18
PDB header: dna binding proteinChain: C: PDB Molecule: dna repair protein rad51;PDBTitle: a crystal structure of the rad51 filament
61 c2zjbB_
not modelled
97.6
13
PDB header: recombinationChain: B: PDB Molecule: meiotic recombination protein dmc1/lim15 homolog;PDBTitle: crystal structure of the human dmc1-m200v polymorphic2 variant
62 c1pznG_
not modelled
97.5
18
PDB header: recombinationChain: G: PDB Molecule: dna repair and recombination protein rad51;PDBTitle: rad51 (rada)
63 c2cvhB_
not modelled
97.4
17
PDB header: dna binding proteinChain: B: PDB Molecule: dna repair and recombination protein radb;PDBTitle: crystal structure of the radb recombinase
64 c3bh0A_
not modelled
97.3
15
PDB header: replicationChain: A: PDB Molecule: dnab-like replicative helicase;PDBTitle: atpase domain of g40p
65 c3cmvG_
not modelled
97.2
18
PDB header: recombinationChain: G: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
66 c2vyeA_
not modelled
97.1
18
PDB header: hydrolaseChain: A: PDB Molecule: replicative dna helicase;PDBTitle: crystal structure of the dnac-ssdna complex
67 c3bs4A_
not modelled
96.8
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ph0321;PDBTitle: crystal structure of uncharacterized protein ph0321 from pyrococcus2 horikoshii in complex with an unknown peptide
68 c1zu4A_
not modelled
96.6
14
PDB header: protein transportChain: A: PDB Molecule: ftsy;PDBTitle: crystal structure of ftsy from mycoplasma mycoides- space2 group p21212
69 d2jdid2
not modelled
96.6
22
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase70 c2q6tB_
not modelled
96.6
27
PDB header: hydrolaseChain: B: PDB Molecule: dnab replication fork helicase;PDBTitle: crystal structure of the thermus aquaticus dnab monomer
71 d1cr2a_
not modelled
96.5
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)72 d1skye2
not modelled
96.5
25
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase73 d1fx0b2
not modelled
96.4
26
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase74 c3bgwD_
not modelled
96.4
23
PDB header: replicationChain: D: PDB Molecule: dnab-like replicative helicase;PDBTitle: the structure of a dnab-like replicative helicase and its interactions2 with primase
75 c2og2A_
not modelled
96.4
18
PDB header: protein transportChain: A: PDB Molecule: putative signal recognition particle receptor;PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
76 c3b9qA_
not modelled
96.4
18
PDB header: protein transportChain: A: PDB Molecule: chloroplast srp receptor homolog, alpha subunitPDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
77 c2j7pA_
not modelled
96.4
16
PDB header: signal recognitionChain: A: PDB Molecule: signal recognition particle protein;PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
78 c3dmdA_
not modelled
96.4
13
PDB header: transport proteinChain: A: PDB Molecule: signal recognition particle receptor;PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
79 d1mabb2
not modelled
96.3
22
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase80 c2cnwF_
not modelled
96.2
20
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
81 c2a5yB_
not modelled
96.1
12
PDB header: apoptosisChain: B: PDB Molecule: ced-4;PDBTitle: structure of a ced-4/ced-9 complex
82 c1vmaA_
not modelled
96.0
21
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
83 c2qy9A_
not modelled
95.8
19
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
84 c3bosA_
not modelled
95.8
14
PDB header: hydrolase regulator,dna binding proteinChain: A: PDB Molecule: putative dna replication factor;PDBTitle: crystal structure of a putative dna replication regulator hda2 (sama_1916) from shewanella amazonensis sb2b at 1.75 a resolution
85 c3dm5A_
not modelled
95.8
17
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
86 d1nlfa_
not modelled
95.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)87 c2iy3A_
not modelled
95.6
16
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein ffh;PDBTitle: structure of the e. coli signal recognition particle2 bound to a translating ribosome
88 c3b85A_
not modelled
95.6
22
PDB header: hydrolaseChain: A: PDB Molecule: phosphate starvation-inducible protein;PDBTitle: crystal structure of predicted phosphate starvation-induced atpase2 phoh2 from corynebacterium glutamicum
89 c2kjqA_
not modelled
95.5
15
PDB header: replicationChain: A: PDB Molecule: dnaa-related protein;PDBTitle: solution structure of protein nmb1076 from neisseria meningitidis.2 northeast structural genomics consortium target mr101b.
90 c2j37W_
not modelled
95.4
13
PDB header: ribosomeChain: W: PDB Molecule: signal recognition particle 54 kda proteinPDBTitle: model of mammalian srp bound to 80s rncs
91 d1vmaa2
not modelled
95.4
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like92 c2yhsA_
not modelled
95.3
15
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the e. coli srp receptor ftsy
93 d1qzxa3
not modelled
95.2
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like94 c1fnnB_
not modelled
95.2
14
PDB header: cell cycleChain: B: PDB Molecule: cell division control protein 6;PDBTitle: crystal structure of cdc6p from pyrobaculum aerophilum
95 c3pvsA_
not modelled
95.1
17
PDB header: recombinationChain: A: PDB Molecule: replication-associated recombination protein a;PDBTitle: structure and biochemical activities of escherichia coli mgsa
96 d1okkd2
not modelled
95.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like97 c2qbyA_
not modelled
94.9
18
PDB header: replication/dnaChain: A: PDB Molecule: cell division control protein 6 homolog 1;PDBTitle: crystal structure of a heterodimer of cdc6/orc1 initiators2 bound to origin dna (from s. solfataricus)
98 c2zamA_
not modelled
94.9
19
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associating protein 4b;PDBTitle: crystal structure of mouse skd1/vps4b apo-form
99 c3j04A_
not modelled
94.7
22
PDB header: structural proteinChain: A: PDB Molecule: myosin-11;PDBTitle: em structure of the heavy meromyosin subfragment of chick smooth2 muscle myosin with regulatory light chain in phosphorylated state
100 c2wjyA_
not modelled
94.6
20
PDB header: hydrolaseChain: A: PDB Molecule: regulator of nonsense transcripts 1;PDBTitle: crystal structure of the complex between human nonsense2 mediated decay factors upf1 and upf2 orthorhombic form
101 d1fnna2
not modelled
94.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain102 c1z6tC_
not modelled
94.3
14
PDB header: apoptosisChain: C: PDB Molecule: apoptotic protease activating factor 1;PDBTitle: structure of the apoptotic protease-activating factor 12 bound to adp
103 c3d8bB_
not modelled
94.2
11
PDB header: hydrolaseChain: B: PDB Molecule: fidgetin-like protein 1;PDBTitle: crystal structure of human fidgetin-like protein 1 in complex with adp
104 c3pfiB_
not modelled
94.1
26
PDB header: hydrolaseChain: B: PDB Molecule: holliday junction atp-dependent dna helicase ruvb;PDBTitle: 2.7 angstrom resolution crystal structure of a probable holliday2 junction dna helicase (ruvb) from campylobacter jejuni subsp. jejuni3 nctc 11168 in complex with adenosine-5'-diphosphate
105 d1qvra2
not modelled
94.0
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain106 d1ls1a2
not modelled
94.0
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like107 d1ye8a1
not modelled
93.9
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)108 c2c9oC_
not modelled
93.9
34
PDB header: hydrolaseChain: C: PDB Molecule: ruvb-like 1;PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
109 c3lxwA_
not modelled
93.9
20
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 1;PDBTitle: crystal structure of human gtpase imap family member 1
110 c3dtpA_
not modelled
93.8
20
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
111 d1nn5a_
not modelled
93.7
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases112 c1xwiA_
not modelled
93.7
16
PDB header: protein transportChain: A: PDB Molecule: skd1 protein;PDBTitle: crystal structure of vps4b
113 d1tmka_
not modelled
93.7
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases114 c1q57G_
not modelled
93.7
7
PDB header: transferaseChain: G: PDB Molecule: dna primase/helicase;PDBTitle: the crystal structure of the bifunctional primase-helicase of2 bacteriophage t7
115 c1nsfA_
not modelled
93.7
20
PDB header: protein transportChain: A: PDB Molecule: n-ethylmaleimide sensitive factor;PDBTitle: d2 hexamerization domain of n-ethylmaleimide sensitive factor (nsf)
116 c3iytG_
not modelled
93.6
21
PDB header: apoptosisChain: G: PDB Molecule: apoptotic protease-activating factor 1;PDBTitle: structure of an apoptosome-procaspase-9 card complex
117 c3e1sA_
not modelled
93.6
17
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease v, subunit recd;PDBTitle: structure of an n-terminal truncation of deinococcus radiodurans recd2
118 c2xzlA_
not modelled
93.5
22
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent helicase nam7;PDBTitle: upf1-rna complex
119 c2ce7B_
not modelled
93.5
16
PDB header: cell division proteinChain: B: PDB Molecule: cell division protein ftsh;PDBTitle: edta treated
120 d1l8qa2
not modelled
93.5
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain