| 1 | c3noyA_
|
|
 |
100.0 |
47 |
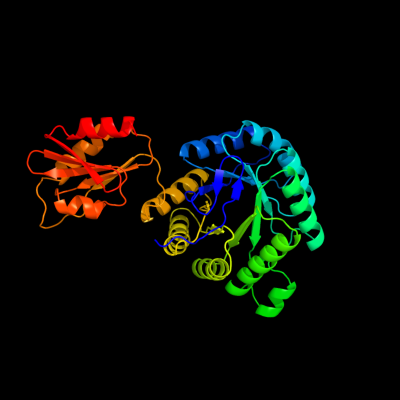
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
| 2 | c2y0fD_
|
|
 |
100.0 |
39 |
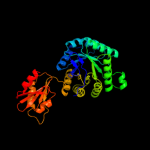
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 3 | c5aopA_
|
|
 |
99.6 |
20 |
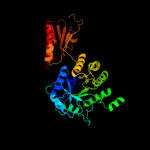
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase hemoprotein;
PDBTitle: sulfite reductase structure reduced with crii edta, 5-coordinate2 siroheme, siroheme feii, [4fe-4s] +1
|
| 4 | c1zj8B_
|
|
 |
99.6 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable ferredoxin-dependent nitrite reductase nira;
PDBTitle: structure of mycobacterium tuberculosis nira protein
|
| 5 | c2akjA_
|
|
 |
99.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin--nitrite reductase, chloroplast;
PDBTitle: structure of spinach nitrite reductase
|
| 6 | d1aopa4
|
|
 |
99.4 |
21 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 7 | c2v4jE_
|
|
 |
99.4 |
21 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 8 | d3c7bb3
|
|
 |
99.4 |
19 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 9 | d1zj8a3
|
|
 |
99.3 |
22 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 10 | d3c7ba3
|
|
 |
99.3 |
21 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 11 | d2akja3
|
|
 |
99.2 |
16 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 12 | d2v4jb3
|
|
 |
99.2 |
19 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 13 | c3c7bE_
|
|
 |
99.2 |
24 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit beta;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 14 | d2v4ja3
|
|
 |
99.2 |
13 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 15 | d2akja4
|
|
 |
99.2 |
18 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 16 | d1aopa3
|
|
 |
99.2 |
12 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 17 | d1tx2a_
|
|
 |
99.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 18 | c1tx2A_
|
|
 |
99.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 19 | c3c7bA_
|
|
 |
99.2 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit alpha;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 20 | d1zj8a4
|
|
 |
99.1 |
16 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 21 | d1ajza_ |
|
not modelled |
99.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 22 | c3tr9A_ |
|
not modelled |
99.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 23 | d1eyea_ |
|
not modelled |
99.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 24 | c2v4jA_ |
|
not modelled |
99.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 25 | c3k13A_ |
|
not modelled |
98.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 26 | c2vp8A_ |
|
not modelled |
98.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
| 27 | c2y5sA_ |
|
not modelled |
98.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 28 | c2yciX_ |
|
not modelled |
98.9 |
18 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 29 | d1f6ya_ |
|
not modelled |
98.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 30 | d3bofa1 |
|
not modelled |
98.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 31 | d1ad1a_ |
|
not modelled |
98.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 32 | c2dzaA_ |
|
not modelled |
98.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 33 | c3bolB_ |
|
not modelled |
98.7 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 34 | c2vefB_ |
|
not modelled |
98.6 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 35 | c3pg8B_ |
|
not modelled |
98.4 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
| 36 | c2h9aA_ |
|
not modelled |
98.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 37 | c1vs1B_ |
|
not modelled |
98.3 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:3-deoxy-7-phosphoheptulonate synthase;
PDBTitle: crystal structure of 3-deoxy-d-arabino-heptulosonate-7-2 phosphate synthase (dahp synthase) from aeropyrum pernix3 in complex with mn2+ and pep
|
| 38 | c3nvtA_ |
|
not modelled |
98.2 |
18 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
| 39 | c2h9aB_ |
|
not modelled |
98.1 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 40 | d1vr6a1 |
|
not modelled |
97.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 41 | c3t4cD_ |
|
not modelled |
97.8 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 1;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
|
| 42 | c3fs2A_ |
|
not modelled |
97.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate2 aldolase from bruciella melitensis at 1.85a resolution
|
| 43 | c2bmbA_ |
|
not modelled |
97.7 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 44 | d2zdra2 |
|
not modelled |
97.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 45 | c2yr1B_ |
|
not modelled |
97.5 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
|
| 46 | c3js3C_ |
|
not modelled |
97.5 |
21 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 47 | c3l2iB_ |
|
not modelled |
97.4 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
| 48 | d1gqna_ |
|
not modelled |
97.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 49 | c1zfjA_ |
|
not modelled |
97.2 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 50 | d1h5ya_ |
|
not modelled |
97.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 51 | c3mcnA_ |
|
not modelled |
97.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
PDBTitle: crystal structure of the 6-hyroxymethyl-7,8-dihydropterin2 pyrophosphokinase dihydropteroate synthase bifunctional enzyme from3 francisella tularensis
|
| 52 | c3q45E_ |
|
not modelled |
97.0 |
17 |
PDB header:isomerase
Chain: E: PDB Molecule:mandelate racemase/muconate lactonizing enzyme family;
PDBTitle: crystal structure of dipeptide epimerase from cytophaga hutchinsonii2 complexed with mg and dipeptide d-ala-l-val
|
| 53 | c2pa6A_ |
|
not modelled |
97.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:enolase;
PDBTitle: crystal structure of mj0232 from methanococcus jannaschii
|
| 54 | c1l8pC_ |
|
not modelled |
97.0 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:enolase 1;
PDBTitle: mg-phosphonoacetohydroxamate complex of s39a yeast enolase 1
|
| 55 | c1zcoA_ |
|
not modelled |
96.9 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphoheptonate aldolase;
PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
|
| 56 | d1o60a_ |
|
not modelled |
96.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 57 | d1jpma1 |
|
not modelled |
96.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 58 | c3q58A_ |
|
not modelled |
96.7 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
| 59 | d2al1a1 |
|
not modelled |
96.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 60 | d1nu5a1 |
|
not modelled |
96.6 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 61 | d1d9ea_ |
|
not modelled |
96.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 62 | c1wueA_ |
|
not modelled |
96.6 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme family
PDBTitle: crystal structure of protein gi:29375081, unknown member of enolase2 superfamily from enterococcus faecalis v583
|
| 63 | c3ijlA_ |
|
not modelled |
96.5 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:muconate cycloisomerase;
PDBTitle: structure of dipeptide epimerase from bacteroides thetaiotaomicron2 complexed with l-pro-d-glu; nonproductive substrate binding.
|
| 64 | c2fkpC_ |
|
not modelled |
96.5 |
22 |
PDB header:isomerase
Chain: C: PDB Molecule:n-acylamino acid racemase;
PDBTitle: the mutant g127c-t313c of deinococcus radiodurans n-2 acylamino acid racemase
|
| 65 | c2zc8B_ |
|
not modelled |
96.5 |
24 |
PDB header:metal binding protein
Chain: B: PDB Molecule:n-acylamino acid racemase;
PDBTitle: crystal structure of n-acylamino acid racemase from thermus2 thermophilus hb8
|
| 66 | d1wbha1 |
|
not modelled |
96.4 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 67 | c2ps2A_ |
|
not modelled |
96.4 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative mandelate racemase/muconate lactonizing
PDBTitle: crystal structure of putative mandelate racemase/muconate2 lactonizing enzyme from aspergillus oryzae
|
| 68 | c3dipA_ |
|
not modelled |
96.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:enolase;
PDBTitle: crystal structure of an enolase protein from the2 environmental genome shotgun sequencing of the sargasso sea
|
| 69 | c2y7eA_ |
|
not modelled |
96.4 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
|
| 70 | c2akmA_ |
|
not modelled |
96.3 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:gamma enolase;
PDBTitle: fluoride inhibition of enolase: crystal structure of the2 inhibitory complex
|
| 71 | c3i4kA_ |
|
not modelled |
96.3 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:muconate lactonizing enzyme;
PDBTitle: crystal structure of muconate lactonizing enzyme from2 corynebacterium glutamicum
|
| 72 | d1rvka1 |
|
not modelled |
96.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 73 | d1r0ma1 |
|
not modelled |
96.3 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 74 | c3qc3B_ |
|
not modelled |
96.3 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
| 75 | d1tqja_ |
|
not modelled |
96.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 76 | d2akza1 |
|
not modelled |
96.3 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 77 | c1jpdX_ |
|
not modelled |
96.3 |
21 |
PDB header:isomerase
Chain: X: PDB Molecule:l-ala-d/l-glu epimerase;
PDBTitle: l-ala-d/l-glu epimerase
|
| 78 | d1pdza1 |
|
not modelled |
96.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 79 | d1yx1a1 |
|
not modelled |
96.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
| 80 | d1jpdx1 |
|
not modelled |
96.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 81 | c3stgA_ |
|
not modelled |
96.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
|
| 82 | c3sz8D_ |
|
not modelled |
96.1 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
| 83 | c1rvkA_ |
|
not modelled |
96.1 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:isomerase/lactonizing enzyme;
PDBTitle: crystal structure of enolase agr_l_2751 from agrobacterium tumefaciens
|
| 84 | c2podA_ |
|
not modelled |
96.1 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:mandelate racemase / muconate lactonizing enzyme;
PDBTitle: crystal structure of a member of enolase superfamily from burkholderia2 pseudomallei k96243
|
| 85 | c3qldB_ |
|
not modelled |
96.1 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:mandelate racemase/muconate lactonizing protein;
PDBTitle: structure of probable mandelate racemase (aalaa1draft_2112) from2 alicyclobacillus acidocaldarius
|
| 86 | c2pgwC_ |
|
not modelled |
96.1 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:muconate cycloisomerase;
PDBTitle: crystal structure of a putative muconate cycloisomerase from2 sinorhizobium meliloti 1021
|
| 87 | c3mqtV_ |
|
not modelled |
96.1 |
13 |
PDB header:isomerase
Chain: V: PDB Molecule:
PDBTitle: crystal structure of a mandelate racemase/muconate lactonizing enzyme2 from shewanella pealeana
|
| 88 | c2qdeA_ |
|
not modelled |
96.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme family
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing family2 protein from azoarcus sp. ebn1
|
| 89 | c1jpmB_ |
|
not modelled |
96.0 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ala-d/l-glu epimerase;
PDBTitle: l-ala-d/l-glu epimerase
|
| 90 | c3ddmD_ |
|
not modelled |
96.0 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:putative mandelate racemase/muconate lactonizing
PDBTitle: crystal structure of mandelate racemase/muconate2 lactonizing enzyme from bordetella bronchiseptica rb50
|
| 91 | c2pozA_ |
|
not modelled |
96.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative dehydratase;
PDBTitle: crystal structure of a putative dehydratase from mesorhizobium loti
|
| 92 | c2p88E_ |
|
not modelled |
96.0 |
19 |
PDB header:lyase
Chain: E: PDB Molecule:mandelate racemase/muconate lactonizing enzyme
PDBTitle: crystal structure of n-succinyl arg/lys racemase from2 bacillus cereus atcc 14579
|
| 93 | c3px5A_ |
|
not modelled |
95.9 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:enzyme of enolase superfamily;
PDBTitle: structure of efi enolase target en500555, a putative dipeptide2 epimerase: apo structure
|
| 94 | c3mkcA_ |
|
not modelled |
95.9 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:racemase;
PDBTitle: crystal structure of a putative racemase
|
| 95 | c3msyC_ |
|
not modelled |
95.8 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from a marine actinobacterium
|
| 96 | c3rcyC_ |
|
not modelled |
95.8 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:mandelate racemase/muconate lactonizing enzyme-like
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme-2 like protein from roseovarius sp. tm1035
|
| 97 | d1vlia2 |
|
not modelled |
95.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 98 | c3igsB_ |
|
not modelled |
95.8 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
| 99 | d2mnra1 |
|
not modelled |
95.8 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 100 | d1ka9f_ |
|
not modelled |
95.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 101 | c2ovlA_ |
|
not modelled |
95.7 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:putative racemase;
PDBTitle: crystal structure of a racemase from streptomyces2 coelicolor a3(2)
|
| 102 | d1muca1 |
|
not modelled |
95.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 103 | c3fv9D_ |
|
not modelled |
95.7 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of putative mandelate racemase/muconatelactonizing2 enzyme from roseovarius nubinhibens ism complexed with magnesium
|
| 104 | c2o56D_ |
|
not modelled |
95.6 |
17 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:putative mandelate racemase;
PDBTitle: crystal structure of a member of the enolase superfamily from2 salmonella typhimurium
|
| 105 | c3dfhC_ |
|
not modelled |
95.6 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:mandelate racemase;
PDBTitle: crystal structure of putative mandelate racemase / muconate2 lactonizing enzyme from vibrionales bacterium swat-3
|
| 106 | c3sjnB_ |
|
not modelled |
95.6 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:mandelate racemase/muconate lactonizing protein;
PDBTitle: crystal structure of enolase spea_3858 (target efi-500646) from2 shewanella pealeana with magnesium bound
|
| 107 | c3dfyJ_ |
|
not modelled |
95.6 |
21 |
PDB header:isomerase
Chain: J: PDB Molecule:muconate cycloisomerase;
PDBTitle: crystal structure of apo dipeptide epimerase from2 thermotoga maritima
|
| 108 | d1h1ya_ |
|
not modelled |
95.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 109 | d1mxsa_ |
|
not modelled |
95.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 110 | d1rpxa_ |
|
not modelled |
95.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 111 | c2c3zA_ |
|
not modelled |
95.5 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
| 112 | c3i6eA_ |
|
not modelled |
95.5 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:muconate cycloisomerase i;
PDBTitle: crystal structure of muconate lactonizing enzyme from2 ruegeria pomeroyi.
|
| 113 | d1sjda1 |
|
not modelled |
95.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 114 | c1sjaA_ |
|
not modelled |
95.4 |
19 |
PDB header:lyase, isomerase
Chain: A: PDB Molecule:n-acylamino acid racemase;
PDBTitle: x-ray structure of o-succinylbenzoate synthase complexed2 with n-acetylmethionine
|
| 115 | c2oqhD_ |
|
not modelled |
95.4 |
13 |
PDB header:isomerase
Chain: D: PDB Molecule:putative isomerase;
PDBTitle: crystal structure of an isomerase from streptomyces coelicolor a3(2)
|
| 116 | d1thfd_ |
|
not modelled |
95.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 117 | c3cb3B_ |
|
not modelled |
95.4 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of l-talarate dehydratase from polaromonas sp. js6662 complexed with mg and l-glucarate
|
| 118 | d1wufa1 |
|
not modelled |
95.3 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 119 | c1nu5A_ |
|
not modelled |
95.3 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:chloromuconate cycloisomerase;
PDBTitle: crystal structure of pseudomonas sp. p51 chloromuconate lactonizing2 enzyme
|
| 120 | d1dvja_ |
|
not modelled |
95.3 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |