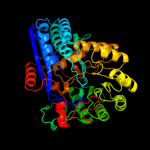
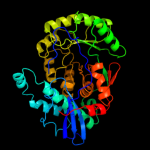
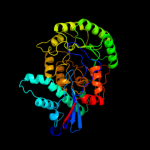
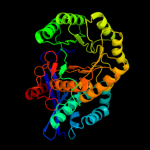
1 c1ec8B_
100.0
100
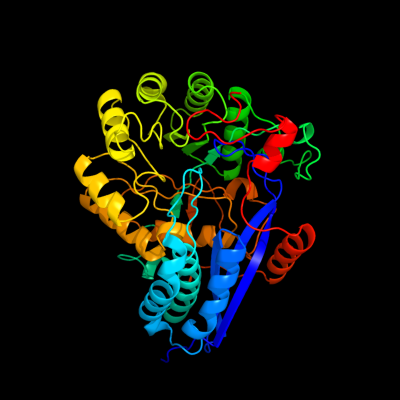
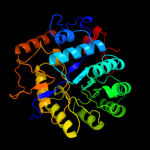
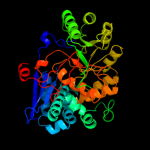
PDB header: lyaseChain: B: PDB Molecule: glucarate dehydratase;PDBTitle: e. coli glucarate dehydratase bound to product 2,3-2 dihydroxy-5-oxo-hexanedioate
2 c3nxlD_
100.0
65
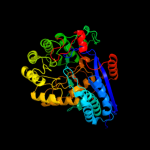
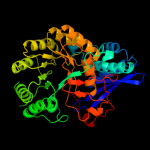
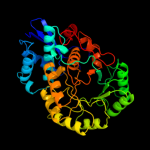
PDB header: lyaseChain: D: PDB Molecule: glucarate dehydratase;PDBTitle: crystal structure of glucarate dehydratase from burkholderia cepacia2 complexed with magnesium
3 c3n6jA_
100.0
57
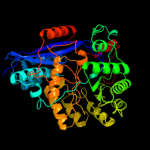
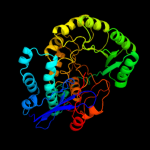
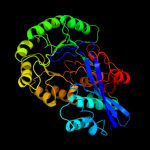
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing protein2 from actinobacillus succinogenes 130z
4 c3mznA_
100.0
59
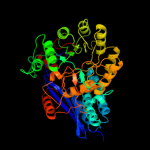
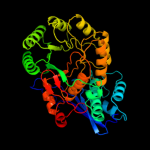
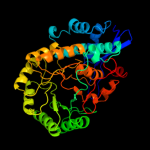
PDB header: lyaseChain: A: PDB Molecule: glucarate dehydratase;PDBTitle: crystal structure of probable glucarate dehydratase from2 chromohalobacter salexigens dsm 3043
5 c2nqlB_
100.0
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: isomerase/lactonizing enzyme;PDBTitle: crystal structure of a member of the enolase superfamily from2 agrobacterium tumefaciens
6 c2pp1C_
100.0
20
PDB header: lyaseChain: C: PDB Molecule: l-talarate/galactarate dehydratase;PDBTitle: crystal structure of l-talarate/galactarate dehydratase from2 salmonella typhimurium lt2 liganded with mg and l-lyxarohydroxamate
7 c2pgwC_
100.0
22
PDB header: isomeraseChain: C: PDB Molecule: muconate cycloisomerase;PDBTitle: crystal structure of a putative muconate cycloisomerase from2 sinorhizobium meliloti 1021
8 d1jdfa1
100.0
100
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like9 c2oo6A_
100.0
21
PDB header: isomeraseChain: A: PDB Molecule: putative l-alanine-dl-glutamate epimerase;PDBTitle: crystal structure of putative l-alanine-dl-glutamate epimerase from2 burkholderia xenovorans strain lb400
10 c3tjiA_
100.0
20
PDB header: lyaseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme, n-terminalPDBTitle: crystal structure of an enolase from enterobacter sp. 638 (efi target2 efi-501662) with bound mg
11 c2qgyA_
100.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: enolase from the environmental genome shotgunPDBTitle: crystal structure of an enolase from the environmental2 genome shotgun sequencing of the sargasso sea
12 c2hxtA_
100.0
15
PDB header: unknown functionChain: A: PDB Molecule: l-fuconate dehydratase;PDBTitle: crystal structure of l-fuconate dehydratase from xanthomonas2 campestris liganded with mg++ and d-erythronohydroxamate
13 c2o56D_
100.0
20
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: putative mandelate racemase;PDBTitle: crystal structure of a member of the enolase superfamily from2 salmonella typhimurium
14 c2chrA_
100.0
20
PDB header: isomeraseChain: A: PDB Molecule: chloromuconate cycloisomerase;PDBTitle: a re-evaluation of the crystal structure of chloromuconate2 cycloisomerase
15 c2ox4E_
100.0
19
PDB header: isomeraseChain: E: PDB Molecule: putative mandelate racemase;PDBTitle: crystal structure of putative dehydratase from zymomonas mobilis zm4
16 c3t6cB_
100.0
21
PDB header: lyaseChain: B: PDB Molecule: putative mand family dehydratase;PDBTitle: crystal structure of an enolase from pantoea ananatis (efi target efi-2 501676) with bound d-gluconate and mg
17 c3rcyC_
100.0
22
PDB header: isomeraseChain: C: PDB Molecule: mandelate racemase/muconate lactonizing enzyme-likePDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme-2 like protein from roseovarius sp. tm1035
18 c2gl5A_
100.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative dehydratase protein;PDBTitle: crystal structure of putative dehydratase from salmonella thyphimurium
19 c3rr1B_
100.0
23
PDB header: lyaseChain: B: PDB Molecule: putative d-galactonate dehydratase;PDBTitle: crystal structure of enolase prk14017 (target efi-500653) from2 ralstonia pickettii 12j
20 c3thuC_
100.0
21
PDB header: lyaseChain: C: PDB Molecule: mandelate racemase / muconate lactonizing enzyme familyPDBTitle: crystal structure of an enolase from sphingomonas sp. ska58 (efi2 target efi-501683) with bound mg
21 c3i6eA_
not modelled
100.0
21
PDB header: isomeraseChain: A: PDB Molecule: muconate cycloisomerase i;PDBTitle: crystal structure of muconate lactonizing enzyme from2 ruegeria pomeroyi.
22 c1nu5A_
not modelled
100.0
21
PDB header: isomeraseChain: A: PDB Molecule: chloromuconate cycloisomerase;PDBTitle: crystal structure of pseudomonas sp. p51 chloromuconate lactonizing2 enzyme
23 c3h12B_
not modelled
100.0
23
PDB header: isomeraseChain: B: PDB Molecule: mandelate racemase;PDBTitle: crystal structure of putative mandelate racemase from bordetella2 bronchiseptica rb50
24 c1rvkA_
not modelled
100.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: isomerase/lactonizing enzyme;PDBTitle: crystal structure of enolase agr_l_2751 from agrobacterium tumefaciens
25 c3dg7B_
not modelled
100.0
23
PDB header: isomeraseChain: B: PDB Molecule: muconate cycloisomerase;PDBTitle: crystal structure of muconate lactonizing enzyme from mucobacterium2 smegmatis complexed with muconolactone
26 c3my9A_
not modelled
100.0
22
PDB header: isomeraseChain: A: PDB Molecule: muconate cycloisomerase;PDBTitle: crystal structure of a muconate cycloisomerase from azorhizobium2 caulinodans
27 c2qddA_
not modelled
100.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of a member of enolase superfamily from roseovarius2 nubinhibens ism
28 c2oqhD_
not modelled
100.0
26
PDB header: isomeraseChain: D: PDB Molecule: putative isomerase;PDBTitle: crystal structure of an isomerase from streptomyces coelicolor a3(2)
29 c2ps2A_
not modelled
100.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative mandelate racemase/muconate lactonizingPDBTitle: crystal structure of putative mandelate racemase/muconate2 lactonizing enzyme from aspergillus oryzae
30 c2qjjC_
not modelled
100.0
20
PDB header: lyaseChain: C: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of d-mannonate dehydratase from novosphingobium2 aromaticivorans
31 c3fv9D_
not modelled
100.0
19
PDB header: hydrolaseChain: D: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of putative mandelate racemase/muconatelactonizing2 enzyme from roseovarius nubinhibens ism complexed with magnesium
32 c1f9cA_
not modelled
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: protein (muconate cycloisomerase i);PDBTitle: crystal structure of mle d178n variant
33 c3i4kA_
not modelled
100.0
22
PDB header: isomeraseChain: A: PDB Molecule: muconate lactonizing enzyme;PDBTitle: crystal structure of muconate lactonizing enzyme from2 corynebacterium glutamicum
34 c3mkcA_
not modelled
100.0
17
PDB header: isomeraseChain: A: PDB Molecule: racemase;PDBTitle: crystal structure of a putative racemase
35 c2qdeA_
not modelled
100.0
21
PDB header: lyaseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme familyPDBTitle: crystal structure of mandelate racemase/muconate lactonizing family2 protein from azoarcus sp. ebn1
36 c3sqsA_
not modelled
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of a putative mandelate racemase/muconate2 lactonizing protein from dinoroseobacter shibae dfl 12
37 c3sjnB_
not modelled
100.0
22
PDB header: lyaseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of enolase spea_3858 (target efi-500646) from2 shewanella pealeana with magnesium bound
38 c3es8D_
not modelled
100.0
19
PDB header: isomerase, lyaseChain: D: PDB Molecule: muconate cycloisomerase;PDBTitle: crystal structure of divergent enolase from oceanobacillus2 iheyensis complexed with mg and l-malate.
39 c3n4eA_
not modelled
100.0
19
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme, c-terminalPDBTitle: crystal structure of mandelate racemase/muconate lactonizing protein2 from paracoccus denitrificans pd1222
40 c3dipA_
not modelled
100.0
20
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of an enolase protein from the2 environmental genome shotgun sequencing of the sargasso sea
41 c3bsmD_
not modelled
100.0
20
PDB header: lyaseChain: D: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of d-mannonate dehydratase from2 chromohalobacter salexigens
42 c2qq6B_
not modelled
100.0
20
PDB header: isomeraseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing enzyme-PDBTitle: crystal structure of mandelate racemase/muconate2 lactonizing enzyme-like protein from rubrobacter3 xylanophilus dsm 9941
43 c3bjsB_
not modelled
100.0
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of a member of enolase superfamily from polaromonas2 sp. js666
44 c3eezA_
not modelled
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: putative mandelate racemase/muconate lactonizingPDBTitle: crystal structure of a putative mandelate racemase/muconate2 lactonizing enzyme from silicibacter pomeroyi
45 c2rdxG_
not modelled
100.0
19
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: mandelate racemase/muconate lactonizing enzyme, putative;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from roseovarius nubinhibens ism
46 c3tj4B_
not modelled
100.0
17
PDB header: lyaseChain: B: PDB Molecule: mandelate racemase;PDBTitle: crystal structure of an enolase from agrobacterium tumefaciens (efi2 target efi-502087) no mg
47 c3q45E_
not modelled
100.0
17
PDB header: isomeraseChain: E: PDB Molecule: mandelate racemase/muconate lactonizing enzyme family;PDBTitle: crystal structure of dipeptide epimerase from cytophaga hutchinsonii2 complexed with mg and dipeptide d-ala-l-val
48 c2gdqB_
not modelled
100.0
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: yitf;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from bacillus subtilis at 1.8 a resolution
49 c2pmqA_
not modelled
100.0
22
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of a mandelate racemase/muconate lactonizing enzyme2 from roseovarius sp. htcc2601
50 c2pozA_
not modelled
100.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative dehydratase;PDBTitle: crystal structure of a putative dehydratase from mesorhizobium loti
51 c3mqtV_
not modelled
100.0
20
PDB header: isomeraseChain: V: PDB Molecule: PDBTitle: crystal structure of a mandelate racemase/muconate lactonizing enzyme2 from shewanella pealeana
52 c2ppgB_
not modelled
100.0
24
PDB header: isomeraseChain: B: PDB Molecule: putative isomerase;PDBTitle: crystal structure of putative isomerase from sinorhizobium meliloti
53 c2podA_
not modelled
100.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mandelate racemase / muconate lactonizing enzyme;PDBTitle: crystal structure of a member of enolase superfamily from burkholderia2 pseudomallei k96243
54 c3t8qA_
not modelled
100.0
21
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme familyPDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 family protein from hoeflea phototrophica
55 c2oz3F_
not modelled
100.0
18
PDB header: lyaseChain: F: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of l-rhamnonate dehydratase from azotobacter2 vinelandii
56 c3dfhC_
not modelled
100.0
17
PDB header: isomeraseChain: C: PDB Molecule: mandelate racemase;PDBTitle: crystal structure of putative mandelate racemase / muconate2 lactonizing enzyme from vibrionales bacterium swat-3
57 c1mraA_
not modelled
100.0
19
PDB header: racemaseChain: A: PDB Molecule: mandelate racemase;PDBTitle: mandelate racemase mutant d270n co-crystallized with (s)-atrolactate
58 c3toyC_
not modelled
100.0
22
PDB header: lyaseChain: C: PDB Molecule: mandelate racemase/muconate lactonizing enzyme familyPDBTitle: crystal structure of enolase brado_4202 (target efi-501651) from2 bradyrhizobium sp. ors278 with calcium and acetate bound
59 c2hzgB_
not modelled
100.0
21
PDB header: isomeraseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing enzyme/enolasePDBTitle: crystal stucture of predicted mandelate racemase from rhodobacter2 sphaeroides
60 c3ugvE_
not modelled
100.0
20
PDB header: lyaseChain: E: PDB Molecule: enolase;PDBTitle: crystal structure of an enolase from alpha pretobacterium bal199 (efi2 target efi-501650) with bound mg
61 c2p88E_
not modelled
100.0
22
PDB header: lyaseChain: E: PDB Molecule: mandelate racemase/muconate lactonizing enzymePDBTitle: crystal structure of n-succinyl arg/lys racemase from2 bacillus cereus atcc 14579
62 c3cxoA_
not modelled
100.0
20
PDB header: lyaseChain: A: PDB Molecule: putative galactonate dehydratase;PDBTitle: crystal structure of l-rhamnonate dehydratase from2 salmonella typhimurium complexed with mg and 3-deoxy-l-3 rhamnonate
63 c3cb3B_
not modelled
100.0
21
PDB header: isomeraseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of l-talarate dehydratase from polaromonas sp. js6662 complexed with mg and l-glucarate
64 c1wueA_
not modelled
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme familyPDBTitle: crystal structure of protein gi:29375081, unknown member of enolase2 superfamily from enterococcus faecalis v583
65 c3t9pB_
not modelled
100.0
23
PDB header: isomeraseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing enzyme familyPDBTitle: crystal structure of a putative mandelate racemase/muconate2 lactonizing enzyme family protein from roseovarius
66 c2ovlA_
not modelled
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: putative racemase;PDBTitle: crystal structure of a racemase from streptomyces2 coelicolor a3(2)
67 c2fkpC_
not modelled
100.0
22
PDB header: isomeraseChain: C: PDB Molecule: n-acylamino acid racemase;PDBTitle: the mutant g127c-t313c of deinococcus radiodurans n-2 acylamino acid racemase
68 c3fcpF_
not modelled
100.0
19
PDB header: isomeraseChain: F: PDB Molecule: l-ala-d/l-glu epimerase, a muconate lactonizingPDBTitle: crystal structure of muconate lactonizing enzyme from2 klebsiella pneumoniae
69 c3cyjA_
not modelled
100.0
21
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme-likePDBTitle: crystal structure of a mandelate racemase/muconate lactonizing enzyme-2 like protein from rubrobacter xylanophilus
70 c3ik4A_
not modelled
100.0
16
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing protein2 from herpetosiphon aurantiacus
71 c2zc8B_
not modelled
100.0
23
PDB header: metal binding proteinChain: B: PDB Molecule: n-acylamino acid racemase;PDBTitle: crystal structure of n-acylamino acid racemase from thermus2 thermophilus hb8
72 c3mwcA_
not modelled
100.0
13
PDB header: ligaseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of probable o-succinylbenzoic acid synthetase from2 kosmotoga olearia
73 c2p0iA_
not modelled
100.0
15
PDB header: lyaseChain: A: PDB Molecule: l-rhamnonate dehydratase;PDBTitle: crystal structure of l-rhamnonate dehydratase from gibberella zeae
74 c1wufB_
not modelled
100.0
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein lin2664;PDBTitle: crystal structure of protein gi:16801725, member of enolase2 superfamily from listeria innocua clip11262
75 c3ddmD_
not modelled
100.0
20
PDB header: lyaseChain: D: PDB Molecule: putative mandelate racemase/muconate lactonizingPDBTitle: crystal structure of mandelate racemase/muconate2 lactonizing enzyme from bordetella bronchiseptica rb50
76 c3n4fD_
not modelled
100.0
20
PDB header: isomeraseChain: D: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing protein2 from geobacillus sp. y412mc10
77 c2dw7G_
not modelled
100.0
16
PDB header: lyaseChain: G: PDB Molecule: bll6730 protein;PDBTitle: crystal structure of d-tartrate dehydratase from2 bradyrhizobium japonicum complexed with mg++ and meso-3 tartrate
78 c1sjaA_
not modelled
100.0
19
PDB header: lyase, isomeraseChain: A: PDB Molecule: n-acylamino acid racemase;PDBTitle: x-ray structure of o-succinylbenzoate synthase complexed2 with n-acetylmethionine
79 c2oz8B_
not modelled
100.0
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: mll7089 protein;PDBTitle: crystal structure of putative mandelate racemase from mesorhizobium2 loti
80 c3msyC_
not modelled
100.0
17
PDB header: isomeraseChain: C: PDB Molecule: mandelate racemase/muconate lactonizing enzyme;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from a marine actinobacterium
81 c3px5A_
not modelled
100.0
15
PDB header: metal binding proteinChain: A: PDB Molecule: enzyme of enolase superfamily;PDBTitle: structure of efi enolase target en500555, a putative dipeptide2 epimerase: apo structure
82 c1jpmB_
not modelled
100.0
21
PDB header: isomeraseChain: B: PDB Molecule: l-ala-d/l-glu epimerase;PDBTitle: l-ala-d/l-glu epimerase
83 c3jw7E_
not modelled
100.0
19
PDB header: isomeraseChain: E: PDB Molecule: dipeptide epimerase;PDBTitle: crystal structure of dipeptide epimerase from enterococcus faecalis2 v583 complexed with mg and dipeptide l-ile-l-tyr
84 c3qldB_
not modelled
100.0
21
PDB header: isomeraseChain: B: PDB Molecule: mandelate racemase/muconate lactonizing protein;PDBTitle: structure of probable mandelate racemase (aalaa1draft_2112) from2 alicyclobacillus acidocaldarius
85 c3ritE_
not modelled
100.0
16
PDB header: isomeraseChain: E: PDB Molecule: dipeptide epimerase;PDBTitle: crystal structure of dipeptide epimerase from methylococcus capsulatus2 complexed with mg and dipeptide l-arg-d-lys
86 c3dfyJ_
not modelled
100.0
20
PDB header: isomeraseChain: J: PDB Molecule: muconate cycloisomerase;PDBTitle: crystal structure of apo dipeptide epimerase from2 thermotoga maritima
87 c3ijlA_
not modelled
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: muconate cycloisomerase;PDBTitle: structure of dipeptide epimerase from bacteroides thetaiotaomicron2 complexed with l-pro-d-glu; nonproductive substrate binding.
88 c2oktA_
not modelled
100.0
16
PDB header: lyaseChain: A: PDB Molecule: o-succinylbenzoic acid synthetase;PDBTitle: crystal structure of o-succinylbenzoic acid synthetase from2 staphylococcus aureus, ligand-free form
89 d1bqga1
not modelled
100.0
74
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like90 c2pgeA_
not modelled
100.0
20
PDB header: lyaseChain: A: PDB Molecule: menc;PDBTitle: crystal structure of menc from desulfotalea psychrophila2 lsv54
91 c1jpdX_
not modelled
100.0
14
PDB header: isomeraseChain: X: PDB Molecule: l-ala-d/l-glu epimerase;PDBTitle: l-ala-d/l-glu epimerase
92 d1muca1
not modelled
100.0
17
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like93 d2gl5a1
not modelled
100.0
18
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like94 d2chra1
not modelled
100.0
18
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like95 d1nu5a1
not modelled
100.0
19
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like96 c3cawB_
not modelled
100.0
16
PDB header: oxidoreductaseChain: B: PDB Molecule: o-succinylbenzoate synthase;PDBTitle: crystal structure of o-succinylbenzoate synthase from2 bdellovibrio bacteriovorus liganded with mg
97 d1rvka1
not modelled
100.0
17
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like98 d1r0ma1
not modelled
100.0
25
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like99 d1wuea1
not modelled
100.0
20
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like100 d2gdqa1
not modelled
100.0
23
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like101 d1wufa1
not modelled
100.0
17
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like102 d1sjda1
not modelled
100.0
24
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like103 d1yeya1
not modelled
100.0
21
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like104 d1tzza1
not modelled
100.0
17
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like105 c2oztA_
not modelled
100.0
17
PDB header: lyaseChain: A: PDB Molecule: tlr1174 protein;PDBTitle: crystal structure of o-succinylbenzoate synthase from2 thermosynechococcus elongatus bp-1
106 d2mnra1
not modelled
100.0
21
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like107 c1kczA_
not modelled
100.0
13
PDB header: lyaseChain: A: PDB Molecule: beta-methylaspartase;PDBTitle: crystal structure of beta-methylaspartase from clostridium2 tetanomorphum. mg-complex.
108 d1jpma1
not modelled
100.0
20
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: D-glucarate dehydratase-like109 c1kkoB_
not modelled
100.0
15
PDB header: lyaseChain: B: PDB Molecule: 3-methylaspartate ammonia-lyase;PDBTitle: crystal structure of citrobacter amalonaticus2 methylaspartate ammonia lyase
110 c2qvhB_
not modelled
100.0
23
PDB header: lyaseChain: B: PDB Molecule: o-succinylbenzoate-coa synthase;PDBTitle: crystal structure of o-succinylbenzoate synthase complexed with o-2 succinyl benzoate (osb)
111 c3gc2A_
not modelled
100.0
21
PDB header: lyaseChain: A: PDB Molecule: o-succinylbenzoate synthase;PDBTitle: 1.85 angstrom crystal structure of o-succinylbenzoate synthase from2 salmonella typhimurium in complex with succinic acid
112 c2pa6A_
not modelled
100.0
14
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of mj0232 from methanococcus jannaschii
113 c3uj2C_
not modelled
100.0
16
PDB header: lyaseChain: C: PDB Molecule: enolase 1;PDBTitle: crystal structure of an enolase from anaerostipes caccae (efi target2 efi-502054) with bound mg and sulfate
114 c3qn3B_
not modelled
100.0
13
PDB header: lyaseChain: B: PDB Molecule: enolase;PDBTitle: phosphopyruvate hydratase from campylobacter jejuni.
115 c1l8pC_
not modelled
100.0
16
PDB header: lyaseChain: C: PDB Molecule: enolase 1;PDBTitle: mg-phosphonoacetohydroxamate complex of s39a yeast enolase 1
116 c3tqpA_
not modelled
100.0
14
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: structure of an enolase (eno) from coxiella burnetii
117 c1iyxA_
not modelled
100.0
16
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of enolase from enterococcus hirae
118 c2akmA_
not modelled
100.0
11
PDB header: lyaseChain: A: PDB Molecule: gamma enolase;PDBTitle: fluoride inhibition of enolase: crystal structure of the2 inhibitory complex
119 c2fymA_
not modelled
100.0
13
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of e. coli enolase complexed with the2 minimal binding segment of rnase e.
120 c2ptwA_
not modelled
100.0
13
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of the t. brucei enolase complexed with2 sulphate, identification of a metal binding site iv