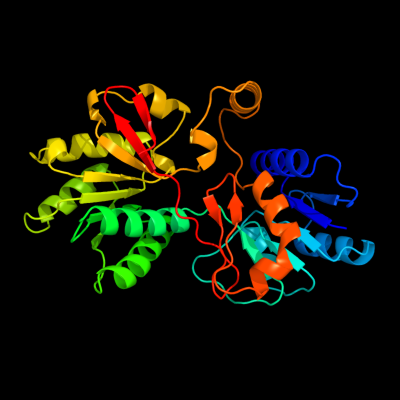
1 d3ckma1
100.0
38
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like2 d1qo0a_
100.0
14
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like3 c3hutA_
100.0
21
PDB header: transport proteinChain: A: PDB Molecule: putative branched-chain amino acid abcPDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from rhodospirillum rubrum
4 c3i09A_
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: periplasmic branched-chain amino acid-binding protein;PDBTitle: crystal structure of a periplasmic binding protein (bma2936) from2 burkholderia mallei at 1.80 a resolution
5 c3t0nA_
100.0
15
PDB header: signaling proteinChain: A: PDB Molecule: twin-arginine translocation pathway signal;PDBTitle: crystal structure of twin-arginine translocation pathway signal from2 rhodopseudomonas palustris bisb5
6 d2liva_
100.0
15
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like7 c3snrA_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: extracellular ligand-binding receptor;PDBTitle: rpd_1889 protein, an extracellular ligand-binding receptor from2 rhodopseudomonas palustris.
8 c3td9A_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: branched chain amino acid abc transporter, periplasmicPDBTitle: crystal structure of a leucine binding protein livk (tm1135) from2 thermotoga maritima msb8 at 1.90 a resolution
9 c3n0wA_
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: abc branched chain amino acid family transporter,PDBTitle: crystal structure of a branched chain amino acid abc transporter2 periplasmic ligand-binding protein (bxe_c0949) from burkholderia3 xenovorans lb400 at 1.88 a resolution
10 c3n0xA_
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: possible substrate binding protein of abc transporterPDBTitle: crystal structure of an abc-type branched-chain amino acid transporter2 (rpa4397) from rhodopseudomonas palustris cga009 at 1.50 a resolution
11 c3lkbB_
100.0
13
PDB header: transport proteinChain: B: PDB Molecule: probable branched-chain amino acid abcPDBTitle: crystal structure of a branched chain amino acid abc2 transporter from thermus thermophilus with bound valine
12 d1usga_
100.0
14
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like13 c3h5lB_
100.0
13
PDB header: transport proteinChain: B: PDB Molecule: putative branched-chain amino acid abcPDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from silicibacter pomeroyi
14 c3i45A_
100.0
14
PDB header: signaling proteinChain: A: PDB Molecule: twin-arginine translocation pathway signal protein;PDBTitle: crystal structure of putative twin-arginine translocation pathway2 signal protein from rhodospirillum rubrum atcc 11170
15 c3ip5A_
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: abc transporter, substrate binding protein (amino acid);PDBTitle: structure of atu2422-gaba receptor in complex with alanine
16 c3sg0A_
100.0
17
PDB header: signaling proteinChain: A: PDB Molecule: extracellular ligand-binding receptor;PDBTitle: the crystal structure of an extracellular ligand-binding receptor from2 rhodopseudomonas palustris haa2
17 c3lopA_
100.0
16
PDB header: substrate binding proteinChain: A: PDB Molecule: substrate binding periplasmic protein;PDBTitle: crystal structure of substrate-binding periplasmic protein2 (pbp) from ralstonia solanacearum
18 c3eafA_
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: abc transporter, substrate binding protein;PDBTitle: crystal structure of abc transporter, substrate binding protein2 aeropyrum pernix
19 d1jdpa_
100.0
13
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like20 c1jdpA_
100.0
13
PDB header: signaling proteinChain: A: PDB Molecule: atrial natriuretic peptide clearance receptor;PDBTitle: crystal structure of hormone/receptor complex
21 c1yk1B_
not modelled
100.0
11
PDB header: hormone/growth factor receptorChain: B: PDB Molecule: atrial natriuretic peptide clearance receptor;PDBTitle: structure of natriuretic peptide receptor-c complexed with brain2 natriuretic peptide
22 c3om1A_
not modelled
100.0
12
PDB header: membrane proteinChain: A: PDB Molecule: glutamate receptor gluk5 (ka2);PDBTitle: crystal structure of the gluk5 (ka2) atd dimer at 1.7 angstrom2 resolution
23 d1dp4a_
not modelled
100.0
13
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like24 c3h6hB_
not modelled
100.0
10
PDB header: membrane proteinChain: B: PDB Molecule: glutamate receptor, ionotropic kainate 2;PDBTitle: crystal structure of the glur6 amino terminal domain dimer assembly2 mpd form
25 c3sm9A_
not modelled
100.0
16
PDB header: signaling proteinChain: A: PDB Molecule: metabotropic glutamate receptor 3;PDBTitle: crystal structure of metabotropic glutamate receptor 3 precursor in2 presence of ly341495 antagonist
26 d1ewka_
not modelled
100.0
12
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like27 c2e4zA_
not modelled
100.0
15
PDB header: signaling proteinChain: A: PDB Molecule: metabotropic glutamate receptor 7;PDBTitle: crystal structure of the ligand-binding region of the group iii2 metabotropic glutamate receptor
28 c3sajB_
not modelled
100.0
14
PDB header: transport proteinChain: B: PDB Molecule: glutamate receptor 1;PDBTitle: crystal structure of glutamate receptor glua1 amino terminal domain
29 c3kg2A_
not modelled
100.0
16
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: glutamate receptor 2;PDBTitle: ampa subtype ionotropic glutamate receptor in complex with competitive2 antagonist zk 200775
30 c3p3wC_
not modelled
100.0
12
PDB header: transport proteinChain: C: PDB Molecule: glutamate receptor 3;PDBTitle: structure of a dimeric glua3 n-terminal domain (ntd) at 4.2 a2 resolution
31 c2e4wA_
not modelled
100.0
17
PDB header: signaling proteinChain: A: PDB Molecule: metabotropic glutamate receptor 3;PDBTitle: crystal structure of the extracellular region of the group ii2 metabotropic glutamate receptor complexed with 1s,3s-acpd
32 c2wjxA_
not modelled
99.9
13
PDB header: transport proteinChain: A: PDB Molecule: glutamate receptor 2;PDBTitle: crystal structure of the human ionotropic glutamate2 receptor glur2 atd region at 4.1 a resolution
33 c3q41B_
not modelled
99.9
18
PDB header: transport proteinChain: B: PDB Molecule: glutamate [nmda] receptor subunit zeta-1;PDBTitle: crystal structure of the glun1 n-terminal domain (ntd)
34 c3jpyA_
not modelled
99.7
14
PDB header: transport proteinChain: A: PDB Molecule: glutamate [nmda] receptor subunit epsilon-2;PDBTitle: crystal structure of the zinc-bound amino terminal domain of the nmda2 receptor subunit nr2b
35 c2iksA_
not modelled
98.6
16
PDB header: transcriptionChain: A: PDB Molecule: dna-binding transcriptional dual regulator;PDBTitle: crystal structure of n-terminal truncated dna-binding transcriptional2 dual regulator from escherichia coli k12
36 c3h5oB_
not modelled
98.5
15
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator gntr;PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
37 c3mizB_
not modelled
98.5
14
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator protein, laciPDBTitle: crystal structure of a putative transcriptional regulator2 protein, lacl family from rhizobium etli
38 c3g85A_
not modelled
98.4
11
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (laci family);PDBTitle: crystal structure of laci family transcription regulator from2 clostridium acetobutylicum
39 c3d8uA_
not modelled
98.4
15
PDB header: transcription regulatorChain: A: PDB Molecule: purr transcriptional regulator;PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
40 c3c3kA_
not modelled
98.4
9
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: crystal structure of an uncharacterized protein from actinobacillus2 succinogenes
41 c3brqA_
not modelled
98.4
10
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional regulator ascg;PDBTitle: crystal structure of the escherichia coli transcriptional repressor2 ascg
42 c3dbiA_
not modelled
98.3
11
PDB header: transcription regulatorChain: A: PDB Molecule: sugar-binding transcriptional regulator, laci family;PDBTitle: crystal structure of sugar-binding transcriptional regulator (laci2 family) from escherichia coli complexed with phosphate
43 c2rjoA_
not modelled
98.3
17
PDB header: signaling proteinChain: A: PDB Molecule: twin-arginine translocation pathway signal protein;PDBTitle: crystal structure of twin-arginine translocation pathway signal2 protein from burkholderia phytofirmans
44 c3gv0A_
not modelled
98.3
19
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
45 c3e3mA_
not modelled
98.3
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of a laci family transcriptional2 regulator from silicibacter pomeroyi
46 c3k4hA_
not modelled
98.3
10
PDB header: transcription regulatorChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator laci from2 bacillus cereus subsp. cytotoxis nvh 391-98
47 c3k9cA_
not modelled
98.3
11
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, laci family protein;PDBTitle: crystal structure of laci transcriptional regulator from rhodococcus2 species.
48 c3qk7C_
not modelled
98.3
15
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulators;PDBTitle: crystal structure of putative transcriptional regulator from yersinia2 pestis biovar microtus str. 91001
49 d1jx6a_
not modelled
98.3
13
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like50 c3hcwB_
not modelled
98.3
9
PDB header: rna binding proteinChain: B: PDB Molecule: maltose operon transcriptional repressor;PDBTitle: crystal structure of probable maltose operon transcriptional repressor2 malr from staphylococcus areus
51 c2rgyA_
not modelled
98.3
12
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of transcriptional regulator of laci family from2 burkhoderia phymatum
52 d1tjya_
not modelled
98.1
14
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like53 c3ksmA_
not modelled
98.1
14
PDB header: transport proteinChain: A: PDB Molecule: abc-type sugar transport system, periplasmic component;PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
54 c3o1hB_
not modelled
98.0
16
PDB header: signaling proteinChain: B: PDB Molecule: periplasmic protein tort;PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
55 d2nzug1
not modelled
98.0
10
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like56 c2qu7B_
not modelled
98.0
10
PDB header: transcriptionChain: B: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of a putative transcription regulator2 from staphylococcus saprophyticus subsp. saprophyticus
57 c3bblA_
not modelled
97.9
9
PDB header: regulatory proteinChain: A: PDB Molecule: regulatory protein of laci family;PDBTitle: crystal structure of a regulatory protein of laci family from2 chloroflexus aggregans
58 c3cs3A_
not modelled
97.9
16
PDB header: transcription regulatorChain: A: PDB Molecule: sugar-binding transcriptional regulator, laci family;PDBTitle: crystal structure of sugar-binding transcriptional regulator (laci2 family) from enterococcus faecalis
59 c3egcF_
not modelled
97.9
15
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative ribose operon repressor;PDBTitle: crystal structure of a putative ribose operon repressor from2 burkholderia thailandensis
60 d1gcaa_
not modelled
97.8
16
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like61 c3h75A_
not modelled
97.8
10
PDB header: sugar binding proteinChain: A: PDB Molecule: periplasmic sugar-binding domain protein;PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
62 c2fqxA_
not modelled
97.6
17
PDB header: transport proteinChain: A: PDB Molecule: membrane lipoprotein tmpc;PDBTitle: pnra from treponema pallidum complexed with guanosine
63 c2ioyB_
not modelled
97.6
17
PDB header: sugar binding proteinChain: B: PDB Molecule: periplasmic sugar-binding protein;PDBTitle: crystal structure of thermoanaerobacter tengcongensis2 ribose binding protein
64 c3lftA_
not modelled
97.6
15
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the abc domain in complex with l-trp from2 streptococcus pneumonia to 1.35a
65 c3jy6B_
not modelled
97.6
19
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of laci transcriptional regulator from lactobacillus2 brevis
66 c2h0aA_
not modelled
97.6
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of probable transcription regulator from2 thermus thermophilus
67 d1jyea_
not modelled
97.5
12
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like68 c1jyeA_
not modelled
97.5
12
PDB header: transcriptionChain: A: PDB Molecule: lactose operon repressor;PDBTitle: structure of a dimeric lac repressor with c-terminal deletion and k84l2 substitution
69 c2o20H_
not modelled
97.5
9
PDB header: transcriptionChain: H: PDB Molecule: catabolite control protein a;PDBTitle: crystal structure of transcription regulator ccpa of lactococcus2 lactis
70 c3ma0A_
not modelled
97.5
18
PDB header: sugar binding proteinChain: A: PDB Molecule: d-xylose-binding periplasmic protein;PDBTitle: closed liganded crystal structure of xylose binding protein from2 escherichia coli
71 d1guda_
not modelled
97.5
15
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like72 c2vk2A_
not modelled
97.5
14
PDB header: transport proteinChain: A: PDB Molecule: abc transporter periplasmic-binding protein ytfq;PDBTitle: crystal structure of a galactofuranose binding protein
73 c3gybB_
not modelled
97.5
14
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulators (laci-familyPDBTitle: crystal structure of a laci-family transcriptional2 regulatory protein from corynebacterium glutamicum
74 c3brsA_
not modelled
97.4
13
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein/laci transcriptional regulator;PDBTitle: crystal structure of sugar transporter from clostridium2 phytofermentans
75 c3kkeA_
not modelled
97.4
15
PDB header: transcription regulatorChain: A: PDB Molecule: laci family transcriptional regulator;PDBTitle: crystal structure of a laci family transcriptional regulator2 from mycobacterium smegmatis
76 c3jvdA_
not modelled
97.3
11
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulators;PDBTitle: crystal structure of putative transcription regulation repressor (laci2 family) from corynebacterium glutamicum
77 c2fn9A_
not modelled
97.3
11
PDB header: sugar binding proteinChain: A: PDB Molecule: ribose abc transporter, periplasmic ribose-binding protein;PDBTitle: thermotoga maritima ribose binding protein unliganded form
78 c3clkB_
not modelled
97.2
6
PDB header: transcription regulatorChain: B: PDB Molecule: transcription regulator;PDBTitle: crystal structure of a transcription regulator from lactobacillus2 plantarum
79 d2dria_
not modelled
97.2
16
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like80 c3d02A_
not modelled
97.2
13
PDB header: sugar binding proteinChain: A: PDB Molecule: putative laci-type transcriptional regulator;PDBTitle: crystal structure of periplasmic sugar-binding protein2 (yp_001338366.1) from klebsiella pneumoniae subsp. pneumoniae mgh3 78578 at 1.30 a resolution
81 c2qvcC_
not modelled
97.1
13
PDB header: transport proteinChain: C: PDB Molecule: sugar abc transporter, periplasmic sugar-bindingPDBTitle: crystal structure of a periplasmic sugar abc transporter2 from thermotoga maritima
82 c1zvvA_
not modelled
97.1
12
PDB header: transcription/dnaChain: A: PDB Molecule: glucose-resistance amylase regulator;PDBTitle: crystal structure of a ccpa-crh-dna complex
83 c3ctpB_
not modelled
97.0
12
PDB header: transcription regulatorChain: B: PDB Molecule: periplasmic binding protein/laci transcriptional regulator;PDBTitle: crystal structure of periplasmic binding protein/laci transcriptional2 regulator from alkaliphilus metalliredigens qymf complexed with d-3 xylulofuranose
84 d8abpa_
not modelled
97.0
8
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like85 c2hqbA_
not modelled
96.8
10
PDB header: transcriptionChain: A: PDB Molecule: transcriptional activator of comk gene;PDBTitle: crystal structure of a transcriptional activator of comk2 gene from bacillus halodurans
86 c2dgdD_
not modelled
96.7
13
PDB header: lyaseChain: D: PDB Molecule: 223aa long hypothetical arylmalonate decarboxylase;PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
87 c2xecD_
not modelled
96.7
13
PDB header: isomeraseChain: D: PDB Molecule: putative maleate isomerase;PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
88 c2qh8A_
not modelled
96.7
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of conserved domain protein from vibrio2 cholerae o1 biovar eltor str. n16961
89 d1byka_
not modelled
96.7
15
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like90 c3bilA_
not modelled
96.6
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable laci-family transcriptional regulator;PDBTitle: crystal structure of a probable laci family transcriptional2 regulator from corynebacterium glutamicum
91 c3g1wB_
not modelled
96.5
7
PDB header: transport proteinChain: B: PDB Molecule: sugar abc transporter;PDBTitle: crystal structure of sugar abc transporter (sugar-binding protein)2 from bacillus halodurans
92 c2x7xA_
not modelled
96.5
8
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: fructose binding periplasmic domain of hybrid two component2 system bt1754
93 d2fvya1
not modelled
96.0
17
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like94 c3l6uA_
not modelled
95.9
10
PDB header: transport proteinChain: A: PDB Molecule: abc-type sugar transport system periplasmicPDBTitle: crystal structure of abc-type sugar transport system,2 periplasmic component from exiguobacterium sibiricum
95 c3s99A_
not modelled
95.5
8
PDB header: lipid binding proteinChain: A: PDB Molecule: basic membrane lipoprotein;PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
96 c2vlbC_
not modelled
95.4
14
PDB header: lyaseChain: C: PDB Molecule: arylmalonate decarboxylase;PDBTitle: structure of unliganded arylmalonate decarboxylase
97 d1dbqa_
not modelled
95.3
14
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like98 c3o74A_
not modelled
95.3
16
PDB header: transcriptionChain: A: PDB Molecule: fructose transport system repressor frur;PDBTitle: crystal structure of cra transcriptional dual regulator from2 pseudomonas putida
99 c3e61A_
not modelled
94.3
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative transcriptional repressor of ribose operon;PDBTitle: crystal structure of a putative transcriptional repressor of ribose2 operon from staphylococcus saprophyticus subsp. saprophyticus
100 c3fflC_
93.2
13
PDB header: cell cycleChain: C: PDB Molecule: anaphase-promoting complex subunit 7;PDBTitle: crystal structure of the n-terminal domain of anaphase-2 promoting complex subunit 7
101 c2xcbA_
93.1
10
PDB header: protein bindingChain: A: PDB Molecule: regulatory protein pcrh;PDBTitle: crystal structure of pcrh in complex with the chaperone2 binding region of popd
102 c2kc7A_
92.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bfr218_protein;PDBTitle: solution nmr structure of bacteroides fragilis protein2 bf1650. northeast structural genomics consortium target3 bfr218
103 d1tlfa_
not modelled
92.5
14
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like104 c3rotA_
not modelled
92.4
11
PDB header: transport proteinChain: A: PDB Molecule: abc sugar transporter, periplasmic sugar binding protein;PDBTitle: crystal structure of abc sugar transporter (periplasmic sugar binding2 protein) from legionella pneumophila
105 c3kjxD_
not modelled
92.3
15
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of a transcriptional regulator, lacl2 family protein from silicibacter pomeroyi
106 c3hs3A_
not modelled
91.7
11
PDB header: transcription regulatorChain: A: PDB Molecule: ribose operon repressor;PDBTitle: crystal structure of periplasmic binding ribose operon2 repressor protein from lactobacillus acidophilus
107 c3k9iA_
91.5
13
PDB header: protein bindingChain: A: PDB Molecule: bh0479 protein;PDBTitle: crystal structure of putative protein binding protein (np_241345.1)2 from bacillus halodurans at 2.71 a resolution
108 c3hymB_
91.1
5
PDB header: cell cycle, ligaseChain: B: PDB Molecule: cell division cycle protein 16 homolog;PDBTitle: insights into anaphase promoting complex tpr subdomain2 assembly from a cdc26-apc6 structure
109 c2r5sB_
91.0
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp0806;PDBTitle: the crystal structure of a domain of protein vp0806 (unknown function)2 from vibrio parahaemolyticus rimd 2210633
110 c2xevB_
not modelled
90.4
12
PDB header: metal bindingChain: B: PDB Molecule: ybgf;PDBTitle: crystal structure of the tpr domain of xanthomonas2 campestris ybgf
111 c3pe3D_
89.3
10
PDB header: transferaseChain: D: PDB Molecule: udp-n-acetylglucosamine--peptide n-PDBTitle: structure of human o-glcnac transferase and its complex with a peptide2 substrate
112 c3l49D_
not modelled
88.2
8
PDB header: transport proteinChain: D: PDB Molecule: abc sugar (ribose) transporter, periplasmicPDBTitle: crystal structure of abc sugar transporter subunit from2 rhodobacter sphaeroides 2.4.1
113 c3gbvB_
not modelled
88.2
17
PDB header: transcription regulatorChain: B: PDB Molecule: putative laci-family transcriptional regulator;PDBTitle: crystal structure of a putative laci transcriptional regulator from2 bacteroides fragilis
114 c3huuC_
not modelled
87.9
11
PDB header: transcription regulatorChain: C: PDB Molecule: transcription regulator like protein;PDBTitle: crystal structure of transcription regulator like protein from2 staphylococcus haemolyticus
115 c2xpiA_
not modelled
87.8
12
PDB header: cell cycleChain: A: PDB Molecule: anaphase-promoting complex subunit cut9;PDBTitle: crystal structure of apc/c hetero-tetramer cut9-hcn1
116 c2dbaA_
not modelled
86.7
13
PDB header: structural proteinChain: A: PDB Molecule: smooth muscle cell associated protein-1, isoformPDBTitle: the solution structure of the tetratrico peptide repeat of2 human smooth muscle cell associated protein-1, isoform 2
117 c3urzB_
not modelled
85.5
11
PDB header: protein bindingChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical protein (bacova_03105) from2 bacteroides ovatus atcc 8483 at 2.19 a resolution
118 d1zbpa1
not modelled
85.2
19
Fold: ImpE-likeSuperfamily: ImpE-likeFamily: ImpE-like119 d1w3ba_
not modelled
85.2
12
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)120 c3mv3B_
not modelled
84.7
11
PDB header: protein transportChain: B: PDB Molecule: coatomer subunit epsilon;PDBTitle: crystal structure of a-cop in complex with e-cop