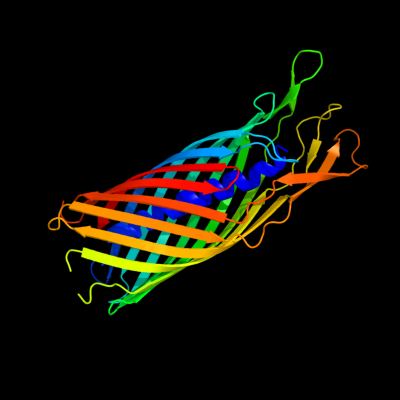
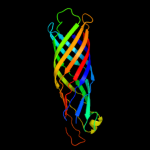
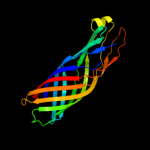
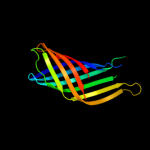
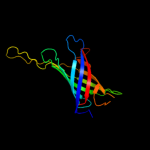
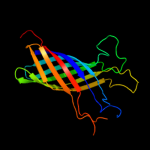
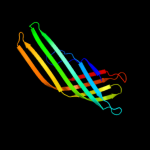
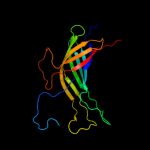
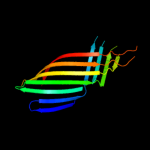
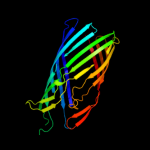
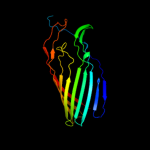
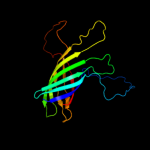
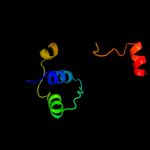
1 c3kvnA_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: esterase esta;PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
2 c3sljA_
100.0
15
PDB header: protein transportChain: A: PDB Molecule: serine protease espp;PDBTitle: pre-cleavage structure of the autotransporter espp - n1023a mutant
3 d1uynx_
99.9
13
Fold: Transmembrane beta-barrelsSuperfamily: AutotransporterFamily: Autotransporter4 c3qq2C_
99.9
18
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
5 c3aehB_
99.9
16
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp autotransporter;PDBTitle: integral membrane domain of autotransporter hbp
6 c2qomB_
99.9
12
PDB header: hydrolaseChain: B: PDB Molecule: serine protease espp;PDBTitle: the crystal structure of the e.coli espp autotransporter beta-domain.
7 c2x27X_
98.0
13
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
8 c3qraA_
97.8
17
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
9 c2f1tB_
97.3
13
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
10 d1qjpa_
97.2
11
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein11 c2jmmA_
97.1
18
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
12 c3nb3C_
96.7
10
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
13 d1p4ta_
96.5
16
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein14 d1g90a_
96.3
13
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein15 d1qj8a_
95.4
12
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein16 c2k0lA_
94.4
11
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
17 d2vdfa1
92.8
15
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane adhesin/invasin OpcA18 d1t16a_
90.7
12
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Outer membrane protein transport protein19 c3dwoX_
90.5
16
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
20 c2lhfA_
88.6
12
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein h1;PDBTitle: solution structure of outer membrane protein h (oprh) from p.2 aeruginosa in dhpc micelles
21 d2zfga1
not modelled
87.5
18
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin22 c2wjqA_
not modelled
86.9
12
PDB header: transport proteinChain: A: PDB Molecule: probable n-acetylneuraminic acid outer membrane channelPDBTitle: nanc porin structure in hexagonal crystal form.
23 c2ervA_
not modelled
83.2
16
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein paer03002360;PDBTitle: crystal structure of the outer membrane enzyme pagl
24 c3dzmB_
not modelled
79.7
15
PDB header: unknown functionChain: B: PDB Molecule: hypothetical conserved protein;PDBTitle: crystal structure of a major outer membrane protein from thermus2 thermophilus hb27
25 c3nsgA_
not modelled
75.0
15
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
26 c3bryB_
not modelled
69.5
11
PDB header: transport proteinChain: B: PDB Molecule: tbux;PDBTitle: crystal structure of the ralstonia pickettii toluene2 transporter tbux
27 d2fgqx1
not modelled
67.8
9
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin28 c2iwvD_
not modelled
65.2
11
PDB header: ion channelChain: D: PDB Molecule: outer membrane protein g;PDBTitle: structure of the monomeric outer membrane porin ompg in the2 open and closed conformation
29 d1vp5a_
64.2
17
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)30 d1osma_
not modelled
64.1
16
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin31 d1c9wa_
not modelled
57.9
26
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)32 d1tlya_
not modelled
54.6
13
Fold: Transmembrane beta-barrelsSuperfamily: Tsx-like channelFamily: Tsx-like channel33 d3eaua1
not modelled
53.9
20
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)34 c2xueB_
not modelled
52.3
23
PDB header: oxidoreductaseChain: B: PDB Molecule: lysine-specific demethylase 6b;PDBTitle: crystal structure of jmjd3
35 c3flpJ_
not modelled
51.6
17
PDB header: sugar binding proteinChain: J: PDB Molecule: sap-like pentraxin;PDBTitle: crystal structure of native heptameric sap-like pentraxin2 from limulus polyphemus
36 c3n6qF_
not modelled
50.8
12
PDB header: oxidoreductaseChain: F: PDB Molecule: yghz aldo-keto reductase;PDBTitle: crystal structure of yghz from e. coli
37 c3n50E_
not modelled
48.9
25
PDB header: transcriptionChain: E: PDB Molecule: transcription factor coe3;PDBTitle: human early b-cell factor 3 (ebf3) ipt/tig and hlhlh domains
38 d1i78a_
not modelled
48.8
10
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane protease OMPT39 d1vqoa1
not modelled
47.6
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L240 c2z4lC_
not modelled
46.4
25
PDB header: ribosomeChain: C: PDB Molecule: 50s ribosomal protein l2;PDBTitle: crystal structure of the bacterial ribosome from escherichia2 coli in complex with paromomycin and ribosome recycling3 factor (rrf). this file contains the 50s subunit of the4 first 70s ribosome, with paromomycin and rrf bound. the5 entire crystal structure contains two 70s ribosomes and is6 described in remark 400.
41 c2k4tA_
not modelled
45.0
13
PDB header: membrane protein,apoptosisChain: A: PDB Molecule: voltage-dependent anion-selective channelPDBTitle: solution structure of human vdac-1 in ldao micelles
42 c3lutA_
not modelled
43.7
20
PDB header: membrane proteinChain: A: PDB Molecule: voltage-gated potassium channel subunit beta-2;PDBTitle: a structural model for the full-length shaker potassium channel kv1.2
43 c3erpA_
not modelled
41.4
14
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxidoreductase;PDBTitle: structure of idp01002, a putative oxidoreductase from and essential2 gene of salmonella typhimurium
44 c2vqiA_
not modelled
41.3
9
PDB header: transportChain: A: PDB Molecule: outer membrane usher protein papc;PDBTitle: structure of the p pilus usher (papc) translocation pore
45 c3up8B_
not modelled
41.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: putative 2,5-diketo-d-gluconic acid reductase b;PDBTitle: crystal structure of a putative 2,5-diketo-d-gluconic acid reductase b
46 d1lqaa_
not modelled
39.6
18
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)47 c3jsbA_
not modelled
38.1
34
PDB header: rna binding proteinChain: A: PDB Molecule: rna-directed rna polymerase;PDBTitle: crystal structure of the n-terminal domain of the lymphocytic2 choriomeningitis virus l protein
48 d1l1fa2
not modelled
36.8
27
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases49 c1ynpA_
not modelled
36.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase;PDBTitle: aldo-keto reductase akr11c1 from bacillus halodurans (apo form)
50 d1pz1a_
not modelled
36.2
16
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)51 d1mi3a_
not modelled
36.1
17
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)52 c2ki0A_
not modelled
35.9
64
PDB header: de novo proteinChain: A: PDB Molecule: ds119;PDBTitle: nmr structure of a de novo designed beta alpha beta
53 c2ke4A_
not modelled
34.9
25
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
54 d1iyjb1
not modelled
34.1
29
Fold: BRCA2 helical domainSuperfamily: BRCA2 helical domainFamily: BRCA2 helical domain55 c3brzA_
not modelled
34.0
12
PDB header: transport proteinChain: A: PDB Molecule: todx;PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
56 c2eo2A_
not modelled
33.6
36
PDB header: oxidoreductaseChain: A: PDB Molecule: adult male hypothalamus cdna, riken full-lengthPDBTitle: solution structure of the insertion region (510-573) of2 fthfs domain from mouse methylenetetrahydrofolate3 dehydrogenase (nadp+ dependent) 1-like protein
57 d2dlqa1
not modelled
32.8
54
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H258 d2j01d1
not modelled
31.6
22
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L259 c1kqsA_
not modelled
30.2
26
PDB header: ribosomeChain: A: PDB Molecule: ribosomal protein l2;PDBTitle: the haloarcula marismortui 50s complexed with a2 pretranslocational intermediate in protein synthesis
60 d1rl2a1
not modelled
29.9
22
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L261 c3a8tA_
not modelled
29.9
17
PDB header: transferaseChain: A: PDB Molecule: adenylate isopentenyltransferase;PDBTitle: plant adenylate isopentenyltransferase in complex with atp
62 c1vliA_
not modelled
29.8
23
PDB header: biosynthetic proteinChain: A: PDB Molecule: spore coat polysaccharide biosynthesis protein spse;PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
63 d1tuza_
not modelled
29.8
27
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins64 c2ns2A_
not modelled
29.7
30
PDB header: cell cycleChain: A: PDB Molecule: spindlin-1;PDBTitle: crystal structure of spindlin1
65 d3proc2
not modelled
29.3
55
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Alpha-lytic protease prodomainFamily: Alpha-lytic protease prodomain66 c3h7rA_
not modelled
29.3
13
PDB header: oxidoreductaseChain: A: PDB Molecule: aldo-keto reductase;PDBTitle: crystal structure of the plant stress-response enzyme akr4c8
67 c2ftcB_
not modelled
29.2
17
PDB header: ribosomeChain: B: PDB Molecule: mitochondrial ribosomal protein l2;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
68 d1vlia2
not modelled
29.2
23
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like69 d1j96a_
not modelled
28.9
19
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)70 d1frba_
not modelled
28.8
23
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)71 d2zjra1
not modelled
28.7
29
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L272 c3buvB_
not modelled
28.5
20
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-oxo-5-beta-steroid 4-dehydrogenase;PDBTitle: crystal structure of human delta(4)-3-ketosteroid 5-beta-reductase in2 complex with nadp and hepes. resolution: 1.35 a.
73 c3njqB_
not modelled
28.5
21
PDB header: viral protein/inhibitorChain: B: PDB Molecule: orf 17;PDBTitle: crystal structure of kaposi's sarcoma-associated herpesvirus protease2 in complex with dimer disruptor
74 c3d5bD_
not modelled
28.5
18
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l2;PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 50s subunit of one 70s ribosome. the entire crystal3 structure contains two 70s ribosomes as described in remark 400.
75 c1tkeA_
not modelled
28.1
29
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: crystal structure of the editing domain of threonyl-trna2 synthetase complexed with serine
76 d1bvua2
not modelled
28.0
30
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases77 c1vbjB_
not modelled
27.7
17
PDB header: oxidoreductaseChain: B: PDB Molecule: prostaglandin f synthase;PDBTitle: the crystal structure of prostaglandin f synthase from2 trypanosoma brucei
78 d1hqta_
not modelled
27.4
21
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)79 c3f7jB_
not modelled
27.4
17
PDB header: oxidoreductaseChain: B: PDB Molecule: yvgn protein;PDBTitle: b.subtilis yvgn
80 c3h7uA_
not modelled
26.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: aldo-keto reductase;PDBTitle: crystal structure of the plant stress-response enzyme akr4c9
81 d1ah4a_
not modelled
26.6
30
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)82 d1b65a_
not modelled
26.4
23
Fold: DmpA/ArgJ-likeSuperfamily: DmpA/ArgJ-likeFamily: DmpA-like83 c2zkra_
not modelled
26.2
23
PDB header: ribosomal protein/rnaChain: A: PDB Molecule: rna expansion segment es3;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
84 d1qr0a1
not modelled
25.9
10
Fold: 4'-phosphopantetheinyl transferaseSuperfamily: 4'-phosphopantetheinyl transferaseFamily: 4'-Phosphopantetheinyl transferase SFP85 c2ze5A_
not modelled
25.7
16
PDB header: transferaseChain: A: PDB Molecule: isopentenyl transferase;PDBTitle: crystal structure of adenosine phosphate-isopentenyltransferase
86 d1afsa_
not modelled
25.6
21
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)87 c2j9uB_
not modelled
25.5
26
PDB header: protein transportChain: B: PDB Molecule: vacuolar protein sorting-associated protein 36;PDBTitle: 2 angstrom x-ray structure of the yeast escrt-i vps28 c-2 terminus in complex with the nzf-n domain from escrt-ii
88 d2j9ub1
not modelled
25.5
26
Fold: Rubredoxin-likeSuperfamily: Ran binding protein zinc finger-likeFamily: Ran binding protein zinc finger-like89 c3b3dA_
not modelled
25.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative morphine dehydrogenase;PDBTitle: b.subtilis ytbe
90 d1ur3m_
not modelled
25.2
12
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)91 d2alra_
not modelled
25.2
24
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)92 c1htoB_
not modelled
25.0
29
PDB header: ligaseChain: B: PDB Molecule: glutamine synthetase;PDBTitle: crystallographic structure of a relaxed glutamine synthetase from2 mycobacterium tuberculosis
93 d1s1pa_
not modelled
24.9
17
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)94 d1miua1
not modelled
24.9
26
Fold: BRCA2 helical domainSuperfamily: BRCA2 helical domainFamily: BRCA2 helical domain95 d1q5ma_
not modelled
24.9
21
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)96 c2yfqA_
not modelled
24.8
40
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-specific glutamate dehydrogenase;PDBTitle: crystal structure of glutamate dehydrogenase from2 peptoniphilus asaccharolyticus
97 d2zdra2
not modelled
24.6
24
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like98 c2gyaA_
not modelled
24.6
17
PDB header: ribosomeChain: A: PDB Molecule: 50s ribosomal protein l2;PDBTitle: structure of the 50s subunit of a pre-translocational e.2 coli ribosome obtained by fitting atomic models for rna and3 protein components into cryo-em map emd-1056
99 c3fozB_
not modelled
24.4
25
PDB header: transferase/rnaChain: B: PDB Molecule: trna delta(2)-isopentenylpyrophosphate transferase;PDBTitle: structure of e. coli isopentenyl-trna transferase in complex with e.2 coli trna(phe)
100 d1gtma2
not modelled
24.3
33
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases101 c4a1cA_
not modelled
24.1
20
PDB header: ribosomeChain: A: PDB Molecule: rpl8;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
102 c2j8pA_
not modelled
23.4
31
PDB header: nuclear proteinChain: A: PDB Molecule: cleavage stimulation factor 64 kda subunit;PDBTitle: nmr structure of c-terminal domain of human cstf-64
103 c3jyzA_
not modelled
23.3
25
PDB header: structural proteinChain: A: PDB Molecule: type iv pilin structural subunit;PDBTitle: crystal structure of pseudomonas aeruginosa (strain:2 pa110594) typeiv pilin in space group p41212
104 c1rl2A_
not modelled
23.2
22
PDB header: ribosomal proteinChain: A: PDB Molecule: protein (ribosomal protein l2);PDBTitle: ribosomal protein l2 rna-binding domain from bacillus2 stearothermophilus
105 d1us0a_
not modelled
22.9
21
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)106 c2b66D_
not modelled
22.8
22
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l2;PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf1,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400
107 c2oz6A_
not modelled
22.7
23
PDB header: dna binding proteinChain: A: PDB Molecule: virulence factor regulator;PDBTitle: crystal structure of virulence factor regulator from pseudomonas2 aeruginosa in complex with camp
108 d2pfxa1
not modelled
22.5
28
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like109 d1qwka_
not modelled
22.4
21
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)110 c3bboE_
not modelled
22.4
33
PDB header: ribosomeChain: E: PDB Molecule: ribosomal protein l2;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
111 c2hb0B_
not modelled
21.8
40
PDB header: cell adhesionChain: B: PDB Molecule: cfa/i fimbrial subunit e;PDBTitle: crystal structure of cfae, the adhesive subunit of cfa/i2 fimbria of enterotoxigenic escherichia coli
112 c2ov3A_
not modelled
21.8
8
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein component of an abcPDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc2 bound
113 d2slia1
not modelled
21.8
36
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Leech intramolecular trans-sialidase, N-terminal domain114 c1kn7A_
not modelled
21.8
42
PDB header: membrane proteinChain: A: PDB Molecule: voltage-gated potassium channel protein kv1.4;PDBTitle: solution structure of the tandem inactivation domain2 (residues 1-75) of potassium channel rck4 (kv1.4)
115 c3ohnA_
not modelled
21.7
7
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane usher protein fimd;PDBTitle: crystal structure of the fimd translocation domain
116 d1u04a1
not modelled
21.7
42
Fold: SH3-like barrelSuperfamily: PAZ domainFamily: PAZ domain117 c1zgdB_
not modelled
21.6
10
PDB header: plant proteinChain: B: PDB Molecule: chalcone reductase;PDBTitle: chalcone reductase complexed with nadp+ at 1.7 angstrom2 resolution
118 c2drhD_
not modelled
21.6
15
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: 361aa long hypothetical d-aminopeptidase;PDBTitle: crystal structure of the ph0078 protein from pyrococcus horikoshii ot3
119 c2b99A_
not modelled
21.5
31
PDB header: transferaseChain: A: PDB Molecule: riboflavin synthase;PDBTitle: crystal structure of an archaeal pentameric riboflavin2 synthase complex with a substrate analog inhibitor
120 c2ql8A_
not modelled
21.3
15
PDB header: oxidoreductaseChain: A: PDB Molecule: putative redox protein;PDBTitle: crystal structure of a putative redox protein (lsei_0423) from2 lactobacillus casei atcc 334 at 1.50 a resolution