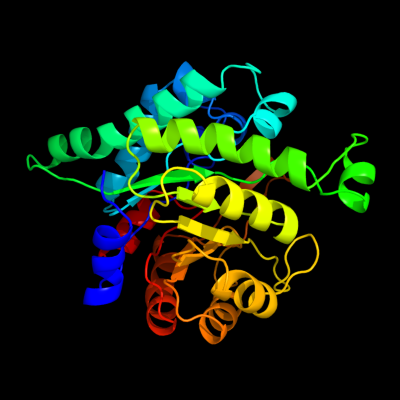
| 1 | d3bofa2
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
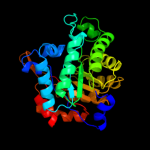
| 2 | c3bolB_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
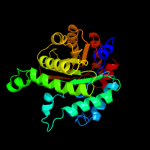
| 3 | d1umya_
|
|
 |
100.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
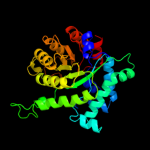
| 4 | d1lt7a_
|
|
 |
100.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 5 | c3eooL_
|
|
 |
94.2 |
17 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 6 | c3lyeA_
|
|
 |
93.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 7 | d1muma_
|
|
 |
93.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 8 | c1zuwA_
|
|
 |
93.4 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase 1;
PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
|
| 9 | c3h5dD_
|
|
 |
91.2 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
| 10 | c2ekcA_
|
|
 |
90.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 11 | d1h1ya_
|
|
 |
89.5 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 12 | d2a6na1
|
|
 |
88.5 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 13 | c3fa4D_
|
|
 |
88.4 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 14 | c3dz1A_
|
|
 |
88.2 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 15 | d1f61a_
|
|
 |
88.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 16 | d1m3ua_
|
|
 |
87.3 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 17 | d1s2wa_
|
|
 |
86.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 18 | d1ujqa_
|
|
 |
86.7 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 19 | c3cprB_
|
|
 |
86.2 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 20 | c2ze3A_
|
|
 |
86.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 21 | d1oy0a_ |
|
not modelled |
84.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 22 | c2dwuA_ |
|
not modelled |
84.6 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
|
| 23 | d1tqxa_ |
|
not modelled |
84.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 24 | c3lciA_ |
|
not modelled |
83.3 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
| 25 | c3ih1A_ |
|
not modelled |
83.2 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 26 | c3kruC_ |
|
not modelled |
82.6 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of the thermostable old yellow enzyme from2 thermoanaerobacter pseudethanolicus e39
|
| 27 | c3s5oA_ |
|
not modelled |
82.5 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 28 | c1jvnB_ |
|
not modelled |
82.4 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 29 | d1gvfa_ |
|
not modelled |
82.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 30 | c2hmcA_ |
|
not modelled |
81.8 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
| 31 | c3g0sA_ |
|
not modelled |
80.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
| 32 | d1vhna_ |
|
not modelled |
79.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 33 | c1zlpA_ |
|
not modelled |
78.1 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 34 | c3lerA_ |
|
not modelled |
77.4 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 35 | c2p10D_ |
|
not modelled |
75.8 |
25 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 36 | d1w3ia_ |
|
not modelled |
75.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 37 | d1pv8a_ |
|
not modelled |
75.7 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 38 | d1o66a_ |
|
not modelled |
74.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 39 | c1zfjA_ |
|
not modelled |
73.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 40 | c3inpA_ |
|
not modelled |
72.8 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
| 41 | c3b8iF_ |
|
not modelled |
72.6 |
23 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 42 | c2r94B_ |
|
not modelled |
71.8 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 43 | c3si9B_ |
|
not modelled |
71.3 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
| 44 | c2vc6A_ |
|
not modelled |
70.3 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 45 | c2gzmB_ |
|
not modelled |
70.3 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of the glutamate racemase from bacillus2 anthracis
|
| 46 | c2y0fD_ |
|
not modelled |
69.7 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 47 | c3hfrA_ |
|
not modelled |
69.5 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
|
| 48 | c2a4aB_ |
|
not modelled |
68.9 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: deoxyribose-phosphate aldolase from p. yoelii
|
| 49 | c3qyqC_ |
|
not modelled |
68.7 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase, putative;
PDBTitle: 1.8 angstrom resolution crystal structure of a putative deoxyribose-2 phosphate aldolase from toxoplasma gondii me49
|
| 50 | c1ps9A_ |
|
not modelled |
68.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 51 | d1a3xa2 |
|
not modelled |
68.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 52 | d1p1xa_ |
|
not modelled |
67.9 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 53 | c3b0vD_ |
|
not modelled |
67.2 |
21 |
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
|
| 54 | d1qopa_ |
|
not modelled |
66.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 55 | c3qz6A_ |
|
not modelled |
66.6 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:hpch/hpai aldolase;
PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
|
| 56 | c2y85D_ |
|
not modelled |
64.3 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:phosphoribosyl isomerase a;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
|
| 57 | d2a4aa1 |
|
not modelled |
63.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 58 | d1pkla2 |
|
not modelled |
62.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 59 | d2flia1 |
|
not modelled |
62.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 60 | d1xcfa_ |
|
not modelled |
62.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 61 | d1ad1a_ |
|
not modelled |
62.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 62 | c3b4uB_ |
|
not modelled |
61.9 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
| 63 | c3js3C_ |
|
not modelled |
61.3 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 64 | c3qc3B_ |
|
not modelled |
60.5 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
| 65 | c3ct7E_ |
|
not modelled |
58.0 |
14 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
| 66 | c3ez4B_ |
|
not modelled |
54.0 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 67 | c2hjpA_ |
|
not modelled |
53.9 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 68 | d16pka_ |
|
not modelled |
53.7 |
30 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 69 | d1ka9f_ |
|
not modelled |
53.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 70 | d1jvna1 |
|
not modelled |
53.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 71 | d1xk7a1 |
|
not modelled |
52.5 |
19 |
Fold:CoA-transferase family III (CaiB/BaiF)
Superfamily:CoA-transferase family III (CaiB/BaiF)
Family:CoA-transferase family III (CaiB/BaiF) |
| 72 | c3fluD_ |
|
not modelled |
52.5 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 73 | d2fdsa1 |
|
not modelled |
51.9 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 74 | c2fdsA_ |
|
not modelled |
51.9 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:orotidine-monophosphate-decarboxylase;
PDBTitle: crystal structure of plasmodium berghei orotidine 5'-2 monophosphate decarboxylase (ortholog of plasmodium3 falciparum pf10_0225)
|
| 75 | c2jfqA_ |
|
not modelled |
51.5 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of staphylococcus aureus glutamate2 racemase in complex with d-glutamate
|
| 76 | c3noeA_ |
|
not modelled |
51.4 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 77 | c2jfoB_ |
|
not modelled |
50.9 |
27 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
|
| 78 | c1vs1B_ |
|
not modelled |
49.8 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:3-deoxy-7-phosphoheptulonate synthase;
PDBTitle: crystal structure of 3-deoxy-d-arabino-heptulosonate-7-2 phosphate synthase (dahp synthase) from aeropyrum pernix3 in complex with mn2+ and pep
|
| 79 | c3ng3A_ |
|
not modelled |
49.6 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of deoxyribose phosphate aldolase from mycobacterium2 avium 104 in a schiff base with an unknown aldehyde
|
| 80 | d1j93a_ |
|
not modelled |
49.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
| 81 | c2ohoA_ |
|
not modelled |
49.2 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: structural basis for glutamate racemase inhibitor
|
| 82 | c3nwrA_ |
|
not modelled |
48.8 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
| 83 | c3navB_ |
|
not modelled |
48.7 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 84 | c3e96B_ |
|
not modelled |
48.7 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 85 | c2jfnA_ |
|
not modelled |
48.4 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
|
| 86 | c3n2xB_ |
|
not modelled |
47.1 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 87 | d1ajza_ |
|
not modelled |
46.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 88 | c1gthD_ |
|
not modelled |
44.8 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydropyrimidine dehydrogenase;
PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
|
| 89 | d1o5ka_ |
|
not modelled |
44.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 90 | d1vzwa1 |
|
not modelled |
44.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 91 | d2a0ma1 |
|
not modelled |
44.0 |
24 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 92 | c1b74A_ |
|
not modelled |
44.0 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase from aquifex pyrophilus
|
| 93 | c3eb2A_ |
|
not modelled |
43.8 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 94 | c3up8B_ |
|
not modelled |
42.2 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative 2,5-diketo-d-gluconic acid reductase b;
PDBTitle: crystal structure of a putative 2,5-diketo-d-gluconic acid reductase b
|
| 95 | c1zcoA_ |
|
not modelled |
41.7 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphoheptonate aldolase;
PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
|
| 96 | c3ff4A_ |
|
not modelled |
41.4 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
| 97 | d2icya2 |
|
not modelled |
40.3 |
24 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 98 | c3daqB_ |
|
not modelled |
40.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 99 | d1dvja_ |
|
not modelled |
39.8 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 100 | c2q4jB_ |
|
not modelled |
39.3 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:probable utp-glucose-1-phosphate uridylyltransferase 2;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at3g03250, a putative udp-glucose3 pyrophosphorylase
|
| 101 | c3tdmD_ |
|
not modelled |
39.1 |
25 |
PDB header:de novo protein
Chain: D: PDB Molecule:computationally designed two-fold symmetric tim-barrel
PDBTitle: computationally designed tim-barrel protein, halfflr
|
| 102 | c2ejaB_ |
|
not modelled |
38.9 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
| 103 | c3l0gD_ |
|
not modelled |
38.6 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
|
| 104 | d1km4a_ |
|
not modelled |
38.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 105 | d1vbga1 |
|
not modelled |
38.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 106 | d1qo2a_ |
|
not modelled |
37.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 107 | d1fw8a_ |
|
not modelled |
37.8 |
21 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 108 | c2ehhE_ |
|
not modelled |
37.7 |
14 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 109 | d1ayia_ |
|
not modelled |
37.7 |
14 |
Fold:Acyl carrier protein-like
Superfamily:Colicin E immunity proteins
Family:Colicin E immunity proteins |
| 110 | d1h5ya_ |
|
not modelled |
37.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 111 | c3qfeB_ |
|
not modelled |
37.4 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
| 112 | c3fokH_ |
|
not modelled |
37.3 |
14 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:uncharacterized protein cgl0159;
PDBTitle: crystal structure of cgl0159 from corynebacterium2 glutamicum (brevibacterium flavum). northeast structural3 genomics target cgr115
|
| 113 | d1xxxa1 |
|
not modelled |
37.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 114 | c2yxgD_ |
|
not modelled |
36.5 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 115 | c1qapA_ |
|
not modelled |
36.5 |
20 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:quinolinic acid phosphoribosyltransferase;
PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
|
| 116 | c1jpkA_ |
|
not modelled |
36.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
|
| 117 | d1twda_ |
|
not modelled |
36.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:CutC-like
Family:CutC-like |
| 118 | d1xkya1 |
|
not modelled |
35.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 119 | d2af4c1 |
|
not modelled |
34.9 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 120 | c2jfzB_ |
|
not modelled |
34.3 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
|