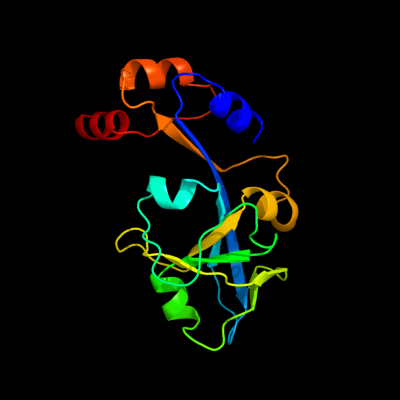
1 c3ftjA_
98.1
16
PDB header: hydrolaseChain: A: PDB Molecule: macrolide export atp-binding/permease proteinPDBTitle: crystal structure of the periplasmic region of macb from2 actinobacillus actinomycetemcomitans
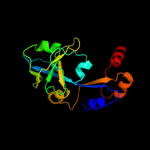
2 c3is6A_
97.9
14
PDB header: transport proteinChain: A: PDB Molecule: putative permease protein, abc transporter;PDBTitle: the crystal structure of a domain of a putative permease protein from2 porphyromonas gingivalis to 2a
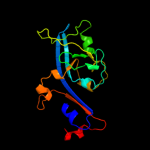
3 c1ciiA_
83.2
14
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
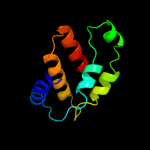
4 d1h0ha1
38.5
15
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain5 d1vlfm1
34.8
18
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain6 d1tmoa1
33.0
18
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain7 d1mvfd_
32.7
16
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: Kis/PemI addiction antidote8 d1kqfa1
30.8
23
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain9 c2iv2X_
29.8
12
PDB header: oxidoreductaseChain: X: PDB Molecule: formate dehydrogenase h;PDBTitle: reinterpretation of reduced form of formate dehydrogenase h2 from e. coli
10 d1ub4c_
29.1
16
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: Kis/PemI addiction antidote11 d1g8ka1
28.9
16
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain12 c2a8vA_
27.3
11
PDB header: protein/rnaChain: A: PDB Molecule: rna binding domain of rho transcriptionPDBTitle: rho transcription termination factor/rna complex
13 d1ogya1
25.9
20
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain14 d2jioa1
25.8
13
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain15 c1eu1A_
25.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: dimethyl sulfoxide reductase;PDBTitle: the crystal structure of rhodobacter sphaeroides dimethylsulfoxide2 reductase reveals two distinct molybdenum coordination environments.
16 d2iv2x1
24.2
13
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain17 c3ltiA_
24.2
38
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase subunit beta;PDBTitle: crystal structure of the escherichia coli rna polymerase beta subunit2 beta2-betai4 domains
18 d1y5ia1
24.0
20
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain19 d1a62a2
22.9
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like20 c2v45A_
22.1
18
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic nitrate reductase;PDBTitle: a new catalytic mechanism of periplasmic nitrate reductase2 from desulfovibrio desulfuricans atcc 27774 from3 crystallographic and epr data and based on detailed4 analysis of the sixth ligand
21 d3dhwa1
not modelled
21.4
15
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like22 d1eu1a1
not modelled
21.0
11
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain23 d1dmra1
not modelled
20.4
11
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain24 c1h5nC_
not modelled
17.6
16
PDB header: oxidoreductaseChain: C: PDB Molecule: dmso reductase;PDBTitle: dmso reductase modified by the presence of dms and air
25 c2ivfA_
not modelled
17.2
21
PDB header: oxidoreductaseChain: A: PDB Molecule: ethylbenzene dehydrogenase alpha-subunit;PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
26 c1h0hA_
not modelled
16.8
15
PDB header: dehydrogenaseChain: A: PDB Molecule: formate dehydrogenase (large subunit);PDBTitle: tungsten containing formate dehydrogenase from2 desulfovibrio gigas
27 c1tmoA_
not modelled
15.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: trimethylamine n-oxide reductase;PDBTitle: trimethylamine n-oxide reductase from shewanella massilia
28 d1e32a1
not modelled
14.3
13
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like29 c2e7zA_
not modelled
13.4
16
PDB header: lyaseChain: A: PDB Molecule: acetylene hydratase ahy;PDBTitle: acetylene hydratase from pelobacter acetylenicus
30 c1vlfQ_
not modelled
12.8
18
PDB header: oxidoreductaseChain: Q: PDB Molecule: pyrogallol hydroxytransferase large subunit;PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici complexed3 with inhibitor 1,2,4,5-tetrahydroxy-benzene
31 c1kqgA_
not modelled
12.7
24
PDB header: oxidoreductaseChain: A: PDB Molecule: formate dehydrogenase, nitrate-inducible, major subunit;PDBTitle: formate dehydrogenase n from e. coli
32 d1cola_
not modelled
12.4
14
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin33 c2yujA_
not modelled
11.9
14
PDB header: protein bindingChain: A: PDB Molecule: ubiquitin fusion degradation 1-like;PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
34 c2pjhB_
not modelled
11.6
13
PDB header: transport proteinChain: B: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: strctural model of the p97 n domain- npl4 ubd complex
35 c2ht2B_
not modelled
11.6
11
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
36 c1g8jC_
not modelled
11.5
17
PDB header: oxidoreductaseChain: C: PDB Molecule: arsenite oxidase;PDBTitle: crystal structure analysis of arsenite oxidase from2 alcaligenes faecalis
37 c2glwA_
not modelled
10.8
23
PDB header: transcriptionChain: A: PDB Molecule: 92aa long hypothetical protein;PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
38 c1zc1A_
not modelled
10.7
14
PDB header: protein turnoverChain: A: PDB Molecule: ubiquitin fusion degradation protein 1;PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
39 c1y5iA_
not modelled
10.7
20
PDB header: oxidoreductaseChain: A: PDB Molecule: respiratory nitrate reductase 1 alpha chain;PDBTitle: the crystal structure of the narghi mutant nari-k86a
40 c1ogyA_
not modelled
10.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic nitrate reductase;PDBTitle: crystal structure of the heterodimeric nitrate reductase2 from rhodobacter sphaeroides
41 d1cz5a1
not modelled
10.4
16
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like42 c3fewX_
not modelled
10.3
17
PDB header: immune systemChain: X: PDB Molecule: colicin s4;PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
43 c2l66B_
not modelled
10.3
8
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, abrb family;PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
44 d1otsa_
not modelled
10.1
12
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel45 c2cwbA_
not modelled
10.1
12
PDB header: protein bindingChain: A: PDB Molecule: chimera of immunoglobulin g binding protein gPDBTitle: solution structure of the ubiquitin-associated domain of2 human bmsc-ubp and its complex with ubiquitin
46 d2vbua1
not modelled
9.9
23
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin kinase-likeFamily: CTP-dependent riboflavin kinase-like47 c1wr1B_
not modelled
9.9
8
PDB header: signaling proteinChain: B: PDB Molecule: ubiquitin-like protein dsk2;PDBTitle: the complex sturcture of dsk2p uba with ubiquitin
48 d1ylea1
not modelled
9.8
31
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: AstA-like49 d2dnaa1
not modelled
9.6
16
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain50 d2bwba1
not modelled
9.5
8
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain51 c2nyaF_
not modelled
9.5
11
PDB header: oxidoreductaseChain: F: PDB Molecule: periplasmic nitrate reductase;PDBTitle: crystal structure of the periplasmic nitrate reductase2 (nap) from escherichia coli
52 c2ki8A_
not modelled
9.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: tungsten formylmethanofuran dehydrogenase,PDBTitle: solution nmr structure of tungsten formylmethanofuran2 dehydrogenase subunit d from archaeoglobus fulgidus,3 northeast structural genomics consortium target att7
53 c2dahA_
not modelled
9.1
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ubiquilin-3;PDBTitle: solution structure of the c-terminal uba domain in the2 human ubiquilin 3
54 c2dnaA_
not modelled
8.8
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: unnamed protein product;PDBTitle: solution structure of rsgi ruh-056, a uba domain from mouse2 cdna
55 d1veja1
not modelled
8.8
24
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain56 d2daha1
not modelled
8.7
16
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain57 c2w1tB_
not modelled
8.7
16
PDB header: transcriptionChain: B: PDB Molecule: stage v sporulation protein t;PDBTitle: crystal structure of b. subtilis spovt
58 d1dzfa2
not modelled
8.6
22
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB559 c2vpyE_
not modelled
8.3
21
PDB header: oxidoreductaseChain: E: PDB Molecule: thiosulfate reductase;PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
60 d2yvxa3
not modelled
8.3
18
Fold: MgtE membrane domain-likeSuperfamily: MgtE membrane domain-likeFamily: MgtE membrane domain-like61 c2fugC_
not modelled
8.3
12
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh-quinone oxidoreductase chain 3;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
62 c2jy5A_
not modelled
7.6
16
PDB header: signaling proteinChain: A: PDB Molecule: ubiquilin-1;PDBTitle: nmr structure of ubiquilin 1 uba domain
63 d1rqpa1
not modelled
7.4
22
Fold: Bacterial fluorinating enzyme, C-terminal domainSuperfamily: Bacterial fluorinating enzyme, C-terminal domainFamily: Bacterial fluorinating enzyme, C-terminal domain64 d1scjb_
not modelled
7.3
16
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors65 c1cz5A_
not modelled
7.2
17
PDB header: hydrolaseChain: A: PDB Molecule: vcp-like atpase;PDBTitle: nmr structure of vat-n: the n-terminal domain of vat (vcp-2 like atpase of thermoplasma)
66 d1hmja_
not modelled
7.2
18
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB567 c2fhdA_
not modelled
7.1
5
PDB header: cell cycleChain: A: PDB Molecule: dna repair protein rhp9/crb2;PDBTitle: crystal structure of crb2 tandem tudor domains
68 d2fy9a1
not modelled
7.1
16
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: AbrB N-terminal domain-like69 d1yfba1
not modelled
7.0
19
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: AbrB N-terminal domain-like70 c1wlfA_
not modelled
6.8
16
PDB header: protein transportChain: A: PDB Molecule: peroxisome biogenesis factor 1;PDBTitle: structure of the n-terminal domain of pex1 aaa-atpase:2 characterization of a putative adaptor-binding domain
71 c2k21A_
not modelled
6.8
11
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
72 c2k29A_
not modelled
6.7
26
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
73 d1dv0a_
not modelled
6.7
8
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain74 c2yvxD_
not modelled
6.6
15
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
75 c3nd0A_
not modelled
6.4
20
PDB header: transport proteinChain: A: PDB Molecule: sll0855 protein;PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
76 c2w2mP_
not modelled
6.3
30
PDB header: hydrolase/receptorChain: P: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: wt pcsk9-deltac bound to wt egf-a of ldlr
77 c2pmwA_
not modelled
6.3
30
PDB header: hydrolaseChain: A: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: the crystal structure of proprotein convertase subtilisin2 kexin type 9 (pcsk9)
78 c2p4eP_
not modelled
6.3
30
PDB header: hydrolaseChain: P: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: crystal structure of pcsk9
79 d2cu3a1
not modelled
6.1
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS80 c2e75E_
not modelled
6.1
20
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
81 c2e76E_
not modelled
6.1
20
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
82 c1vf5R_
not modelled
6.1
20
PDB header: photosynthesisChain: R: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
83 d2e74e1
not modelled
6.1
20
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex84 c1vf5E_
not modelled
6.1
20
PDB header: photosynthesisChain: E: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
85 c2e74E_
not modelled
6.1
20
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
86 c2kncB_
not modelled
6.0
12
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
87 d1spbp_
not modelled
6.0
15
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors88 c2kncA_
not modelled
5.9
22
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
89 d1yloa1
not modelled
5.7
24
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain90 d1zq1a1
not modelled
5.7
11
Fold: Sm-like foldSuperfamily: GatD N-terminal domain-likeFamily: GatD N-terminal domain-like91 d1q08a_
not modelled
5.7
8
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators92 c2kl0A_
not modelled
5.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
93 c2cw5B_
not modelled
5.6
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: bacterial fluorinating enzyme homolog;PDBTitle: crystal structure of a conserved hypothetical protein from2 thermus thermophilus hb8
94 c2jpcA_
not modelled
5.6
23
PDB header: dna binding proteinChain: A: PDB Molecule: ssrb;PDBTitle: ssrb dna binding protein
95 d1dgsa2
not modelled
5.6
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain96 d2d6fa1
not modelled
5.5
14
Fold: Sm-like foldSuperfamily: GatD N-terminal domain-likeFamily: GatD N-terminal domain-like97 d3d31c1
not modelled
5.5
15
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like98 c3d31D_
not modelled
5.5
15
PDB header: transport proteinChain: D: PDB Molecule: sulfate/molybdate abc transporter, permeasePDBTitle: modbc from methanosarcina acetivorans
99 c2f4nA_
not modelled
5.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein mj1651;PDBTitle: crystal structure of protein mj1651 from methanococcus2 jannaschii dsm 2661, pfam duf62