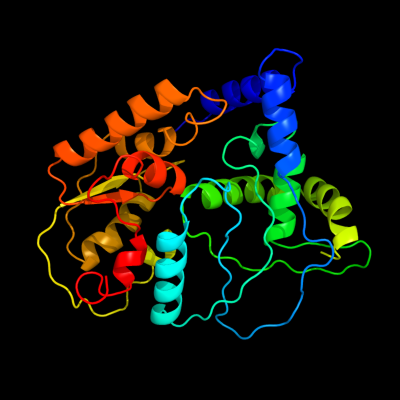
1 c3kwlA_
99.7
10
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical protein from helicobacter pylori
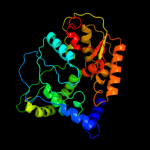
2 c2h89B_
99.6
23
PDB header: oxidoreductaseChain: B: PDB Molecule: succinate dehydrogenase ip subunit;PDBTitle: avian respiratory complex ii with malonate bound
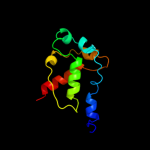
3 c1nekB_
99.5
21
PDB header: oxidoreductase/electron transportChain: B: PDB Molecule: succinate dehydrogenase iron-sulfur protein;PDBTitle: complex ii (succinate dehydrogenase) from e. coli with2 ubiquinone bound
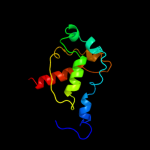
4 d1nekb1
99.5
21
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain5 c2b76N_
99.5
19
PDB header: oxidoreductaseChain: N: PDB Molecule: fumarate reductase iron-sulfur protein;PDBTitle: e. coli quinol fumarate reductase frda e49q mutation
6 d1kf6b1
99.5
20
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain7 c2bs2E_
99.4
18
PDB header: oxidoreductaseChain: E: PDB Molecule: quinol-fumarate reductase iron-sulfur subunit b;PDBTitle: quinol:fumarate reductase from wolinella succinogenes
8 d2bs2b1
99.4
19
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain9 c3cf4A_
99.1
17
PDB header: oxidoreductaseChain: A: PDB Molecule: acetyl-coa decarboxylase/synthase alpha subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex
10 c1gx7A_
98.2
17
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic [fe] hydrogenase large subunit;PDBTitle: best model of the electron transfer complex between2 cytochrome c3 and [fe]-hydrogenase
11 c1hfeL_
98.1
17
PDB header: hydrogenaseChain: L: PDB Molecule: protein (fe-only hydrogenase (e.c.1.18.99.1)PDBTitle: 1.6 a resolution structure of the fe-only hydrogenase from2 desulfovibrio desulfuricans
12 c1c4cA_
98.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (fe-only hydrogenase);PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
13 d2c42a5
97.6
22
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins14 d1jb0c_
97.4
31
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin15 d2fug91
97.4
28
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins16 c2fugG_
97.4
28
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase chain 9;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
17 d3c8ya3
97.3
19
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins18 c2c3yA_
97.3
38
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate-ferredoxin oxidoreductase;PDBTitle: crystal structure of the radical form of2 pyruvate:ferredoxin oxidoreductase from desulfovibrio3 africanus
19 d1xera_
97.1
20
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Archaeal ferredoxins20 c2vdcI_
97.0
15
PDB header: oxidoreductaseChain: I: PDB Molecule: glutamate synthase [nadph] small chain;PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
21 d1gtea5
not modelled
97.0
29
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins22 c1gthD_
not modelled
97.0
35
PDB header: oxidoreductaseChain: D: PDB Molecule: dihydropyrimidine dehydrogenase;PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
23 c2gmhA_
not modelled
96.9
23
PDB header: oxidoreductaseChain: A: PDB Molecule: electron transfer flavoprotein-ubiquinonePDBTitle: structure of porcine electron transfer flavoprotein-2 ubiquinone oxidoreductase in complexed with ubiquinone
24 d1hfel2
not modelled
96.8
30
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins25 d2fug34
not modelled
96.8
22
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins26 d1rgva_
not modelled
96.8
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins27 c1kqfB_
not modelled
96.8
32
PDB header: oxidoreductaseChain: B: PDB Molecule: formate dehydrogenase, nitrate-inducible, iron-sulfurPDBTitle: formate dehydrogenase n from e. coli
28 d1blua_
not modelled
96.7
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins29 d2fdna_
not modelled
96.7
33
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins30 c2fgoA_
not modelled
96.6
20
PDB header: electron transportChain: A: PDB Molecule: ferredoxin;PDBTitle: structure of the 2[4fe-4s] ferredoxin from pseudomonas2 aeruginosa
31 d7fd1a_
not modelled
96.5
36
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin32 d1bc6a_
not modelled
96.5
31
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin33 d1dura_
not modelled
96.5
32
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins34 d1clfa_
not modelled
96.4
40
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins35 c2zvsB_
not modelled
96.3
31
PDB header: electron transportChain: B: PDB Molecule: uncharacterized ferredoxin-like protein yfhl;PDBTitle: crystal structure of the 2[4fe-4s] ferredoxin from escherichia coli
36 d1fcaa_
not modelled
96.3
31
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins37 c2ivfB_
not modelled
96.3
43
PDB header: oxidoreductaseChain: B: PDB Molecule: ethylbenzene dehydrogenase beta-subunit;PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
38 d1y5ib1
not modelled
96.3
43
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins39 c2fugC_
not modelled
96.1
24
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh-quinone oxidoreductase chain 3;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
40 d1h98a_
not modelled
96.0
29
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin41 c3gyxJ_
not modelled
95.9
33
PDB header: oxidoreductaseChain: J: PDB Molecule: adenylylsulfate reductase;PDBTitle: crystal structure of adenylylsulfate reductase from2 desulfovibrio gigas
42 d1kqfb1
not modelled
95.8
32
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins43 d1h0hb_
not modelled
95.8
16
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins44 d2gmha3
not modelled
95.8
22
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: ETF-QO domain-like45 d1jnrb_
not modelled
95.7
29
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins46 c1ti2F_
not modelled
95.5
27
PDB header: oxidoreductaseChain: F: PDB Molecule: pyrogallol hydroxytransferase small subunit;PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici
47 d1vlfn2
not modelled
95.4
27
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins48 c3c7bE_
not modelled
95.1
31
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunit beta;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
49 c2vpyB_
not modelled
95.0
30
PDB header: oxidoreductaseChain: B: PDB Molecule: nrfc protein;PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
50 d1vjwa_
not modelled
94.7
30
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin51 c2v2kB_
not modelled
94.7
34
PDB header: electron transportChain: B: PDB Molecule: ferredoxin;PDBTitle: the crystal structure of fdxa, a 7fe ferredoxin from2 mycobacterium smegmatis
52 c1dwlA_
not modelled
94.5
21
PDB header: electron transferChain: A: PDB Molecule: ferredoxin i;PDBTitle: the ferredoxin-cytochrome complex using heteronuclear nmr2 and docking simulation
53 c2v4jE_
not modelled
94.1
26
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
54 d1iqza_
not modelled
94.0
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin55 d1sj1a_
not modelled
93.4
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin56 c2fugA_
not modelled
92.8
14
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase chain 1;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
57 d3c7bb1
not modelled
92.5
32
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins58 d1gtea1
not modelled
91.0
20
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Dihydropyrimidine dehydrogenase, N-terminal domain59 c3c7bA_
not modelled
90.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunit alpha;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
60 d1fxra_
not modelled
89.4
17
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin61 d1jfla1
not modelled
85.2
14
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase62 c3bk7A_
not modelled
83.5
28
PDB header: hydrolyase/translationChain: A: PDB Molecule: abc transporter atp-binding protein;PDBTitle: structure of the complete abce1/rnaase-l inhibitor protein2 from pyrococcus abysii
63 c2v4jA_
not modelled
83.2
23
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
64 c2zskA_
not modelled
79.7
18
PDB header: unknown functionChain: A: PDB Molecule: 226aa long hypothetical aspartate racemase;PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
65 c2jfzB_
not modelled
79.2
15
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
66 c3hfrA_
not modelled
78.9
16
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
67 c2ohoA_
not modelled
76.3
17
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: structural basis for glutamate racemase inhibitor
68 d2v4jb1
not modelled
73.7
32
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins69 c2jfoB_
not modelled
73.0
16
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
70 c1zuwA_
not modelled
72.8
7
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase 1;PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
71 c3outC_
not modelled
70.9
10
PDB header: isomeraseChain: C: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
72 c3ojcD_
not modelled
70.9
18
PDB header: isomeraseChain: D: PDB Molecule: putative aspartate/glutamate racemase;PDBTitle: crystal structure of a putative asp/glu racemase from yersinia pestis
73 c2dwuA_
not modelled
70.7
12
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
74 c2jfqA_
not modelled
68.7
17
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of staphylococcus aureus glutamate2 racemase in complex with d-glutamate
75 c2jfnA_
not modelled
67.6
12
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
76 c1b74A_
not modelled
66.8
15
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: glutamate racemase from aquifex pyrophilus
77 c3gjzB_
not modelled
66.3
13
PDB header: immune systemChain: B: PDB Molecule: microcin immunity protein mccf;PDBTitle: crystal structure of microcin immunity protein mccf from bacillus2 anthracis str. ames
78 c2yv4A_
not modelled
62.3
16
PDB header: rna binding proteinChain: A: PDB Molecule: hypothetical protein ph0435;PDBTitle: crystal structure of c-terminal sua5 domain from pyrococcus horikoshii2 hypothetical sua5 protein ph0435
79 c2vdcF_
not modelled
57.6
53
PDB header: oxidoreductaseChain: F: PDB Molecule: glutamate synthase [nadph] large chain;PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
80 c2gzmB_
not modelled
56.0
12
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of the glutamate racemase from bacillus2 anthracis
81 c1lm1A_
not modelled
55.2
53
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin-dependent glutamate synthase;PDBTitle: structural studies on the synchronization of catalytic centers in2 glutamate synthase: native enzyme
82 c2dx7B_
not modelled
55.2
16
PDB header: isomeraseChain: B: PDB Molecule: aspartate racemase;PDBTitle: crystal structure of pyrococcus horikoshii ot3 aspartate racemase2 complex with citric acid
83 c1zrsB_
not modelled
54.0
15
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protein;PDBTitle: wild-type ld-carboxypeptidase
84 d2auna2
not modelled
52.4
15
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like85 d1ofda2
not modelled
50.2
53
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases86 c2vlbC_
not modelled
45.6
11
PDB header: lyaseChain: C: PDB Molecule: arylmalonate decarboxylase;PDBTitle: structure of unliganded arylmalonate decarboxylase
87 d2fzva1
not modelled
42.4
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase88 c1g8jC_
not modelled
41.0
33
PDB header: oxidoreductaseChain: C: PDB Molecule: arsenite oxidase;PDBTitle: crystal structure analysis of arsenite oxidase from2 alcaligenes faecalis
89 c3l4eA_
not modelled
40.0
15
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized peptidase lmo0363;PDBTitle: 1.5a crystal structure of a putative peptidase e protein from listeria2 monocytogenes egd-e
90 d2dx7a1
not modelled
39.6
14
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase91 c3lrmB_
not modelled
36.5
8
PDB header: hydrolaseChain: B: PDB Molecule: alpha-galactosidase 1;PDBTitle: structure of alfa-galactosidase from saccharomyces cerevisiae with2 raffinose
92 d1zl0a2
not modelled
36.1
14
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like93 d1ea0a2
not modelled
35.3
53
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases94 d2c4ka2
not modelled
31.3
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like95 c3s81A_
not modelled
25.7
11
PDB header: isomeraseChain: A: PDB Molecule: putative aspartate racemase;PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
96 d1b74a2
not modelled
25.2
16
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase97 d1i60a_
not modelled
24.8
10
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like98 d1lxja_
not modelled
23.9
4
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like99 c3rotA_
not modelled
22.7
12
PDB header: transport proteinChain: A: PDB Molecule: abc sugar transporter, periplasmic sugar binding protein;PDBTitle: crystal structure of abc sugar transporter (periplasmic sugar binding2 protein) from legionella pneumophila
100 d1ei9a_
not modelled
21.6
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterases101 c2kvgA_
not modelled
20.8
42
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein