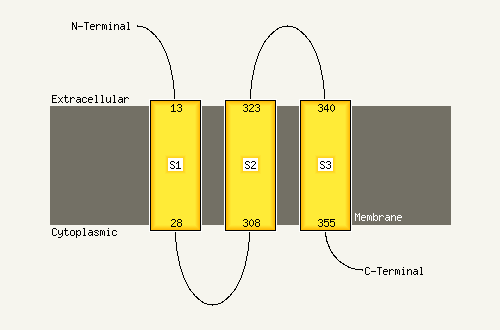
1 c3ojaB_
97.4
13
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
2 c1bf5A_
96.9
7
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: tyrosine phosphorylated stat-1/dna complex
3 c1c1gA_
96.9
8
PDB header: contractile proteinChain: A: PDB Molecule: tropomyosin;PDBTitle: crystal structure of tropomyosin at 7 angstroms resolution2 in the spermine-induced crystal form
4 c1ciiA_
96.8
7
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
5 c1deqF_
96.3
7
PDB header: PDB COMPND: 6 c1bg1A_
96.2
6
PDB header: transcription/dnaChain: A: PDB Molecule: protein (transcription factor stat3b);PDBTitle: transcription factor stat3b/dna complex
7 c3cwgA_
95.7
7
PDB header: transcriptionChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: unphosphorylated mouse stat3 core fragment
8 c1yvlB_
95.4
7
PDB header: signaling proteinChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: structure of unphosphorylated stat1
9 c2efrB_
95.4
9
PDB header: contractile proteinChain: B: PDB Molecule: general control protein gcn4 and tropomyosin 1 alpha chain;PDBTitle: crystal structure of the c-terminal tropomyosin fragment with n- and2 c-terminal extensions of the leucine zipper at 1.8 angstroms3 resolution
10 c3o0zD_
94.5
11
PDB header: transferaseChain: D: PDB Molecule: rho-associated protein kinase 1;PDBTitle: crystal structure of a coiled-coil domain from human rock i
11 c1ei3E_
94.3
10
PDB header: PDB COMPND: 12 c3ghgK_
93.7
12
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
13 c2d3eD_
92.9
9
PDB header: contractile proteinChain: D: PDB Molecule: general control protein gcn4 and tropomyosin 1PDBTitle: crystal structure of the c-terminal fragment of rabbit2 skeletal alpha-tropomyosin
14 c3na7A_
92.6
8
PDB header: gene regulation, chaperoneChain: A: PDB Molecule: hp0958;PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
15 c2v71A_
92.5
9
PDB header: nuclear proteinChain: A: PDB Molecule: nuclear distribution protein nude-like 1;PDBTitle: coiled-coil region of nudel
16 c1jchC_
92.4
3
PDB header: ribosome inhibitor, hydrolaseChain: C: PDB Molecule: colicin e3;PDBTitle: crystal structure of colicin e3 in complex with its immunity protein
17 c2v66C_
92.2
4
PDB header: structural proteinChain: C: PDB Molecule: nuclear distribution protein nude-like 1;PDBTitle: crystal structure of the coiled-coil domain of ndel1 (a.a.2 58 to 169)c
18 c4a55B_
92.0
7
PDB header: transferaseChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit alpha;PDBTitle: crystal structure of p110alpha in complex with ish2 of p85alpha and2 the inhibitor pik-108
19 c2fxmB_
91.2
7
PDB header: contractile proteinChain: B: PDB Molecule: myosin heavy chain, cardiac muscle beta isoform;PDBTitle: structure of the human beta-myosin s2 fragment
20 c2gl2B_
90.7
15
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
21 c1deqO_
not modelled
90.5
11
PDB header: PDB COMPND: 22 c1ei3C_
not modelled
89.2
6
PDB header: PDB COMPND: 23 c3u59C_
not modelled
89.0
17
PDB header: contractile proteinChain: C: PDB Molecule: tropomyosin beta chain;PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
24 c1deqD_
not modelled
87.4
11
PDB header: PDB COMPND: 25 c3ol1A_
not modelled
86.7
6
PDB header: structural proteinChain: A: PDB Molecule: vimentin;PDBTitle: crystal structure of vimentin (fragment 144-251) from homo sapiens,2 northeast structural genomics consortium target hr4796b
26 c2b9cA_
not modelled
86.2
11
PDB header: contractile proteinChain: A: PDB Molecule: striated-muscle alpha tropomyosin;PDBTitle: structure of tropomyosin's mid-region: bending and binding2 sites for actin
27 c3hnwB_
85.5
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
28 c3hizB_
not modelled
77.6
6
PDB header: transferase/oncoproteinChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunitPDBTitle: crystal structure of p110alpha h1047r mutant in complex with2 nish2 of p85alpha
29 c2rd0B_
not modelled
72.3
8
PDB header: transferase/oncoproteinChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit alpha;PDBTitle: structure of a human p110alpha/p85alpha complex
30 c2i1jA_
not modelled
70.9
11
PDB header: cell adhesion, membrane proteinChain: A: PDB Molecule: moesin;PDBTitle: moesin from spodoptera frugiperda at 2.1 angstroms resolution
31 c2v1yB_
not modelled
62.0
9
PDB header: transferaseChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit alpha;PDBTitle: structure of a phosphoinositide 3-kinase alpha adaptor-2 binding domain (abd) in a complex with the ish2 domain3 from p85 alpha
32 c3ojaA_
not modelled
61.6
7
PDB header: protein bindingChain: A: PDB Molecule: leucine-rich immune molecule 1;PDBTitle: crystal structure of lrim1/apl1c complex
33 c1gk4A_
not modelled
54.1
10
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment (cys2)
34 c2no2A_
not modelled
51.9
8
PDB header: cell adhesionChain: A: PDB Molecule: huntingtin-interacting protein 1;PDBTitle: crystal structure of the dllrkn-containing coiled-coil2 domain of huntingtin-interacting protein 1
35 d2pila_
not modelled
49.8
13
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin36 c3dtpA_
not modelled
48.5
7
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
37 c3ipkA_
not modelled
47.3
8
PDB header: cell adhesionChain: A: PDB Molecule: agi/ii;PDBTitle: crystal structure of a3vp1 of agi/ii of streptococcus mutans
38 c1hciB_
not modelled
45.2
12
PDB header: triple-helix coiled coilChain: B: PDB Molecule: alpha-actinin 2;PDBTitle: crystal structure of the rod domain of alpha-actinin
39 c2wpqA_
not modelled
41.8
11
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
40 c3sokB_
not modelled
38.8
20
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
41 c2voyB_
not modelled
38.4
16
PDB header: hydrolaseChain: B: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
42 c1y4cA_
not modelled
36.8
15
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
43 c1sjjB_
not modelled
32.9
8
PDB header: contractile proteinChain: B: PDB Molecule: actinin;PDBTitle: cryo-em structure of chicken gizzard smooth muscle alpha-2 actinin
44 c1wt6B_
not modelled
32.6
21
PDB header: transferaseChain: B: PDB Molecule: myotonin-protein kinase;PDBTitle: coiled-coil domain of dmpk
45 d1oqwa_
not modelled
32.5
18
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin46 c2e7sM_
not modelled
32.2
6
PDB header: endocytosis/exocytosisChain: M: PDB Molecule: rab guanine nucleotide exchange factor sec2;PDBTitle: crystal structure of the yeast sec2p gef domain
47 c2jeeA_
not modelled
31.5
6
PDB header: cell cycleChain: A: PDB Molecule: yiiu;PDBTitle: xray structure of e. coli yiiu
48 d2ap3a1
not modelled
31.4
10
Fold: Four-helical up-and-down bundleSuperfamily: MW0975(SA0943)-likeFamily: MW0975(SA0943)-like49 c2y3aB_
not modelled
28.3
5
PDB header: transferaseChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunit beta;PDBTitle: crystal structure of p110beta in complex with icsh2 of p85beta and2 the drug gdc-0941
50 c2dq3A_
not modelled
26.5
14
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of aq_298
51 c3u1aC_
not modelled
23.4
14
PDB header: contractile proteinChain: C: PDB Molecule: smooth muscle tropomyosin alpha;PDBTitle: n-terminal 81-aa fragment of smooth muscle tropomyosin alpha
52 c3q0xA_
not modelled
22.4
18
PDB header: structural proteinChain: A: PDB Molecule: centriole protein;PDBTitle: n-terminal coiled-coil dimer domain of c. reinhardtii sas-6 homolog2 bld12p
53 c3mraA_
not modelled
21.1
16
PDB header: membrane proteinChain: A: PDB Molecule: nicotinic acetylcholine receptor;PDBTitle: m3 transmembrane segment of alpha-subunit of nicotinic2 acetylcholine receptor from torpedo californica, nmr, 153 structures
54 c3ajwA_
not modelled
19.1
4
PDB header: protein transportChain: A: PDB Molecule: flagellar flij protein;PDBTitle: structure of flij, a soluble component of flagellar type iii export2 apparatus
55 c1m1jA_
not modelled
18.4
7
PDB header: blood clottingChain: A: PDB Molecule: fibrinogen alpha subunit;PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
56 c2oevA_
not modelled
18.3
9
PDB header: protein transportChain: A: PDB Molecule: programmed cell death 6-interacting protein;PDBTitle: crystal structure of alix/aip1
57 c2qa7B_
not modelled
16.7
5
PDB header: actin bindingChain: B: PDB Molecule: huntingtin-interacting protein 1;PDBTitle: crystal structure of huntingtin-interacting protein 12 (hip1) coiled-coil domain with a basic surface suitable3 for hip-protein interactor (hippi)
58 d1gl4a1
not modelled
16.6
50
Fold: GFP-likeSuperfamily: GFP-likeFamily: Domain G2 of nidogen-159 c1h4uA_
not modelled
15.6
50
PDB header: extracellular matrix proteinChain: A: PDB Molecule: nidogen-1;PDBTitle: domain g2 of mouse nidogen-1
60 d1jb0m_
not modelled
15.4
32
Fold: Single transmembrane helixSuperfamily: Subunit XII of photosystem I reaction centre, PsaMFamily: Subunit XII of photosystem I reaction centre, PsaM61 c1jb0M_
not modelled
15.4
32
PDB header: photosynthesisChain: M: PDB Molecule: photosystem 1 reaction centre subunit xii;PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
62 d2axti1
not modelled
15.3
11
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like63 c3tnfB_
not modelled
14.8
9
PDB header: protein transportChain: B: PDB Molecule: lida;PDBTitle: lida from legionella in complex with active rab8a
64 c2jo1A_
not modelled
13.7
13
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
65 c1x8yA_
not modelled
13.5
13
PDB header: structural proteinChain: A: PDB Molecule: lamin a/c;PDBTitle: human lamin coil 2b
66 c3a7pB_
not modelled
13.1
12
PDB header: protein transportChain: B: PDB Molecule: autophagy protein 16;PDBTitle: the crystal structure of saccharomyces cerevisiae atg16
67 c3cvfA_
not modelled
12.4
9
PDB header: signaling proteinChain: A: PDB Molecule: homer protein homolog 3;PDBTitle: crystal structure of the carboxy terminus of homer3
68 d1seta1
not modelled
12.2
17
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Seryl-tRNA synthetase (SerRS)69 c2jp3A_
not modelled
11.7
13
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
70 c3mk7B_
not modelled
11.6
17
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit o;PDBTitle: the structure of cbb3 cytochrome oxidase
71 c2oexB_
not modelled
11.4
9
PDB header: protein transportChain: B: PDB Molecule: programmed cell death 6-interacting protein;PDBTitle: structure of alix/aip1 v domain
72 c1l8dB_
not modelled
11.4
9
PDB header: replicationChain: B: PDB Molecule: dna double-strand break repair rad50 atpase;PDBTitle: rad50 coiled-coil zn hook
73 c1afoB_
not modelled
11.3
11
PDB header: integral membrane proteinChain: B: PDB Molecule: glycophorin a;PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
74 d1q90m_
not modelled
11.3
25
Fold: Single transmembrane helixSuperfamily: PetM subunit of the cytochrome b6f complexFamily: PetM subunit of the cytochrome b6f complex75 c2jwaA_
not modelled
10.6
22
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
76 c2wvrB_
not modelled
9.9
17
PDB header: replicationChain: B: PDB Molecule: geminin;PDBTitle: human cdt1:geminin complex
77 c2dq0A_
not modelled
9.5
9
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of seryl-trna synthetase from pyrococcus2 horikoshii complexed with a seryl-adenylate analog
78 c3m9bK_
not modelled
9.2
22
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
79 c3qh9A_
not modelled
9.0
11
PDB header: structural proteinChain: A: PDB Molecule: liprin-beta-2;PDBTitle: human liprin-beta2 coiled-coil
80 c1debA_
not modelled
8.7
17
PDB header: structural proteinChain: A: PDB Molecule: adenomatous polyposis coli protein;PDBTitle: crystal structure of the n-terminal coiled coil domain from2 apc
81 c2zxeG_
not modelled
8.5
31
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
82 c2xdjF_
not modelled
7.8
11
PDB header: unknown functionChain: F: PDB Molecule: uncharacterized protein ybgf;PDBTitle: crystal structure of the n-terminal domain of e.coli ybgf
83 c3pcqM_
not modelled
7.6
32
PDB header: photosynthesisChain: M: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: femtosecond x-ray protein nanocrystallography
84 c2ap7A_
not modelled
7.6
37
PDB header: antibioticChain: A: PDB Molecule: bombinin h2;PDBTitle: solution structure of bombinin h2 in dpc micelles
85 c2eqbC_
not modelled
7.3
13
PDB header: endocytosis/exocytosisChain: C: PDB Molecule: rab guanine nucleotide exchange factor sec2;PDBTitle: crystal structure of the rab gtpase sec4p, the sec2p gef2 domain, and phosphate complex
86 c2hacA_
not modelled
7.2
17
PDB header: membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd3 zeta chain;PDBTitle: structure of zeta-zeta transmembrane dimer
87 c3l9oA_
not modelled
7.1
7
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: crystal structure of mtr4, a co-factor of the nuclear exosome
88 c3movB_
not modelled
7.1
10
PDB header: structural proteinChain: B: PDB Molecule: lamin-b1;PDBTitle: crystal structure of human lamin-b1 coil 2 segment
89 c3n4xB_
not modelled
7.0
10
PDB header: replicationChain: B: PDB Molecule: monopolin complex subunit csm1;PDBTitle: structure of csm1 full-length
90 c1fosF_
not modelled
7.0
14
PDB header: transcription/dnaChain: F: PDB Molecule: c-jun proto-oncogene protein;PDBTitle: two human c-fos:c-jun:dna complexes
91 c3n23E_
not modelled
6.6
25
PDB header: hydrolaseChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
92 c3lssA_
not modelled
6.5
12
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: trypanosoma brucei seryl-trna synthetase in complex with atp
93 c2zxxA_
not modelled
6.1
17
PDB header: cell cycle/replicationChain: A: PDB Molecule: geminin;PDBTitle: crystal structure of cdt1/geminin complex
94 c3pdsA_
not modelled
6.1
4
PDB header: membrane protein/hydrolaseChain: A: PDB Molecule: fusion protein beta-2 adrenergic receptor/lysozyme;PDBTitle: irreversible agonist-beta2 adrenoceptor complex
95 c1ic2B_
not modelled
5.9
18
PDB header: contractile proteinChain: B: PDB Molecule: tropomyosin alpha chain, skeletal muscle;PDBTitle: deciphering the design of the tropomyosin molecule
96 c1f5nA_
not modelled
5.8
7
PDB header: signaling proteinChain: A: PDB Molecule: interferon-induced guanylate-binding protein 1;PDBTitle: human guanylate binding protein-1 in complex with the gtp2 analogue, gmppnp.
97 d1v54i_
not modelled
5.6
22
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIcFamily: Mitochondrial cytochrome c oxidase subunit VIc98 c1pi8A_
not modelled
5.5
25
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: structure of the channel-forming trans-membrane domain of2 virus protein "u" (vpu) from hiv-1
99 c2gohA_
not modelled
5.5
25
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: three-dimensional structure of the trans-membrane domain of2 vpu from hiv-1 in aligned phospholipid bicelles