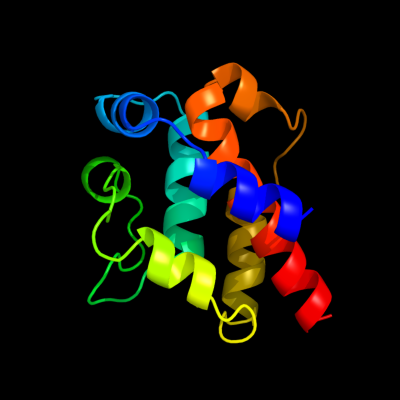
| 1 | c3j1uA_
|
|
 |
98.7 |
14 |
PDB header:motor protein/structural protein
Chain: A: PDB Molecule:cytoplasmic dynein 1 heavy chain 1, seryl t-rna synthetase
PDBTitle: low affinity dynein microtubule binding domain - tubulin complex
|
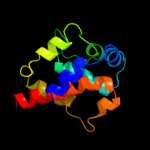
| 2 | c2rr7A_
|
|
 |
98.5 |
14 |
PDB header:motor protein
Chain: A: PDB Molecule:dynein heavy chain 9;
PDBTitle: microtubule binding domain of dynein-c
|
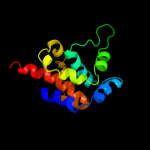
| 3 | c3errB_
|
|
 |
98.3 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:fusion protein of microtubule binding domain from
PDBTitle: microtubule binding domain from mouse cytoplasmic dynein as2 a fusion with seryl-trna synthetase
|
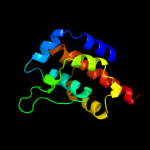
| 4 | c3ibjB_
|
|
 |
98.0 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:cgmp-dependent 3',5'-cyclic phosphodiesterase;
PDBTitle: x-ray structure of pde2a
|
| 5 | c3mf0A_
|
|
 |
97.8 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:cgmp-specific 3',5'-cyclic phosphodiesterase;
PDBTitle: crystal structure of pde5a gaf domain (89-518)
|
| 6 | c3vkhA_
|
|
 |
97.7 |
16 |
PDB header:motor protein
Chain: A: PDB Molecule:dynein heavy chain, cytoplasmic;
PDBTitle: x-ray structure of a functional full-length dynein motor domain
|
| 7 | c1mc0A_
|
|
 |
97.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:3',5'-cyclic nucleotide phosphodiesterase 2a;
PDBTitle: regulatory segment of mouse 3',5'-cyclic nucleotide phosphodiesterase2 2a, containing the gaf a and gaf b domains
|
| 8 | c3o4xE_
|
|
 |
97.3 |
15 |
PDB header:protein binding
Chain: E: PDB Molecule:protein diaphanous homolog 1;
PDBTitle: crystal structure of complex between amino and carboxy terminal2 fragments of mdia1
|
| 9 | d1ux5a_
|
|
 |
97.2 |
13 |
Fold:Formin homology 2 domain (FH2 domain)
Superfamily:Formin homology 2 domain (FH2 domain)
Family:Formin homology 2 domain (FH2 domain) |
| 10 | c3o4xF_
|
|
 |
97.2 |
14 |
PDB header:protein binding
Chain: F: PDB Molecule:protein diaphanous homolog 1;
PDBTitle: crystal structure of complex between amino and carboxy terminal2 fragments of mdia1
|
| 11 | c1y64B_
|
|
 |
96.9 |
13 |
PDB header:structural protein
Chain: B: PDB Molecule:bni1 protein;
PDBTitle: bni1p formin homology 2 domain complexed with atp-actin
|
| 12 | c2lb5A_
|
|
 |
96.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:sensor histidine kinase;
PDBTitle: refined structural basis for the photoconversion of a phytochrome to2 the activated far-red light-absorbing form
|
| 13 | d1f5ma_
|
|
 |
96.8 |
20 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 14 | d1v9da_
|
|
 |
96.7 |
16 |
Fold:Formin homology 2 domain (FH2 domain)
Superfamily:Formin homology 2 domain (FH2 domain)
Family:Formin homology 2 domain (FH2 domain) |
| 15 | c1vhmB_
|
|
 |
96.7 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein yebr;
PDBTitle: crystal structure of an hypothetical protein
|
| 16 | c3oovA_
|
|
 |
96.7 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:methyl-accepting chemotaxis protein, putative;
PDBTitle: crystal structure of a methyl-accepting chemotaxis protein, residues2 122 to 287
|
| 17 | c3p01C_
|
|
 |
96.7 |
13 |
PDB header:signaling protein
Chain: C: PDB Molecule:two-component response regulator;
PDBTitle: crystal structure of two-component response regulator from nostoc sp.2 pcc 7120
|
| 18 | c4fofA_
|
|
 |
96.6 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:methyl-accepting chemotaxis protein;
PDBTitle: crystal structure of the blue-light absorbing form of the2 thermosynechococcus elongatus pixj gaf-domain
|
| 19 | d1vhma_
|
|
 |
96.4 |
19 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 20 | c2vjwA_
|
|
 |
96.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:gaf family protein;
PDBTitle: crystal structure of the second gaf domain of devs from2 mycobacterium smegmatis
|
| 21 | c3rfbB_ |
|
not modelled |
96.3 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of frmsr
|
| 22 | c1ykdB_ |
|
not modelled |
96.3 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:adenylate cyclase;
PDBTitle: crystal structure of the tandem gaf domains from a cyanobacterial2 adenylyl cyclase: novel modes of ligand-binding and dimerization
|
| 23 | c3ksiA_ |
|
not modelled |
96.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of frmsr of staphylococcus aureus (complex with 2-propanol)
|
| 24 | c3p01A_ |
|
not modelled |
96.3 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:two-component response regulator;
PDBTitle: crystal structure of two-component response regulator from nostoc sp.2 pcc 7120
|
| 25 | c4g3kB_ |
|
not modelled |
96.2 |
25 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator nlh1;
PDBTitle: crystal structure of a. aeolicus nlh1 gaf domain in an inactive state
|
| 26 | c3mmhA_ |
|
not modelled |
96.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase;
PDBTitle: x-ray structure of free methionine-r-sulfoxide reductase from2 neisseria meningitidis in complex with its substrate
|
| 27 | c4g3vB_ |
|
not modelled |
96.2 |
16 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator nlh2;
PDBTitle: crystal structure of a. aeolicus nlh2 gaf domain in an inactive state
|
| 28 | c3eeaB_ |
|
not modelled |
96.0 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:gaf domain/hd domain protein;
PDBTitle: the crystal structure of the gaf domain/hd domain protein2 from geobacter sulfurreducens
|
| 29 | c2k31A_ |
|
not modelled |
95.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphodiesterase 5a, cgmp-specific;
PDBTitle: solution structure of cgmp-binding gaf domain of2 phosphodiesterase 5
|
| 30 | c4eahE_ |
|
not modelled |
95.9 |
7 |
PDB header:protein binding
Chain: E: PDB Molecule:formin-like protein 3;
PDBTitle: crystal structure of the formin homology 2 domain of fmnl3 bound to2 actin
|
| 31 | d2o9ca1 |
|
not modelled |
95.9 |
11 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 32 | c2qybA_ |
|
not modelled |
95.8 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:membrane protein, putative;
PDBTitle: crystal structure of the gaf domain region of putative membrane2 protein from geobacter sulfurreducens pca
|
| 33 | c3k2nB_ |
|
not modelled |
95.7 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:sigma-54-dependent transcriptional regulator;
PDBTitle: the crystal structure of sigma-54-dependent transcriptional2 regulator domain from chlorobium tepidum tls
|
| 34 | c2j1dG_ |
|
not modelled |
95.5 |
9 |
PDB header:protein binding
Chain: G: PDB Molecule:disheveled-associated activator of morphogenesis 1;
PDBTitle: crystallization of hdaam1 c-terminal fragment
|
| 35 | c3dbaB_ |
|
not modelled |
95.4 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:cone cgmp-specific 3',5'-cyclic phosphodiesterase subunit
PDBTitle: crystal structure of the cgmp-bound gaf a domain from the2 photoreceptor phosphodiesterase 6c
|
| 36 | c2z6eC_ |
|
not modelled |
95.4 |
11 |
PDB header:protein fibril regulator
Chain: C: PDB Molecule:disheveled-associated activator of morphogenesis
PDBTitle: crystal structure of human daam1 fh2
|
| 37 | d2k2na1 |
|
not modelled |
95.4 |
16 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 38 | c3trcA_ |
|
not modelled |
95.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the gaf domain from a phosphoenolpyruvate-protein2 phosphotransferase (ptsp) from coxiella burnetii
|
| 39 | c2w3gA_ |
|
not modelled |
95.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:two component sensor histidine kinase devs (gaf
PDBTitle: air-oxidized structure of the first gaf domain of2 mycobacterium tuberculosis doss
|
| 40 | c3ci6B_ |
|
not modelled |
95.1 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: crystal structure of the gaf domain from acinetobacter2 phosphoenolpyruvate-protein phosphotransferase
|
| 41 | c3hcyB_ |
|
not modelled |
94.5 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative two-component sensor histidine kinase protein;
PDBTitle: the crystal structure of the domain of putative two-component sensor2 histidine kinase protein from sinorhizobium meliloti 1021
|
| 42 | d2veaa1 |
|
not modelled |
94.4 |
16 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 43 | c3o5yA_ |
|
not modelled |
94.3 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:sensor protein;
PDBTitle: the crystal structure of the gaf domain of a two-component sensor2 histidine kinase from bacillus halodurans to 2.45a
|
| 44 | c3w2zA_ |
|
not modelled |
94.1 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:methyl-accepting chemotaxis protein;
PDBTitle: crystal structure of the cyanobacterial protein
|
| 45 | d1p0za_ |
|
not modelled |
93.2 |
18 |
Fold:Profilin-like
Superfamily:Sensory domain-like
Family:Sensory domain of two-component sensor kinase |
| 46 | c2zmfA_ |
|
not modelled |
93.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:camp and camp-inhibited cgmp 3',5'-cyclic phosphodiesterase
PDBTitle: crystal structure of the c-terminal gaf domain of human2 phosphodiesterase 10a
|
| 47 | c2oolA_ |
|
not modelled |
90.9 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of the chromophore-binding domain of an unusual2 bacteriophytochrome rpbphp3 from r. palustris
|
| 48 | c3e0yA_ |
|
not modelled |
88.9 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of a conserved domain from a protein of2 geobacter sulfurreducens pca
|
| 49 | d1mc0a1 |
|
not modelled |
88.6 |
10 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 50 | d1mc0a2 |
|
not modelled |
88.0 |
14 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 51 | d2oola1 |
|
not modelled |
87.4 |
8 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 52 | d3c2wa1 |
|
not modelled |
86.6 |
8 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
| 53 | c3e98B_ |
|
not modelled |
86.0 |
12 |
PDB header:unknown function
Chain: B: PDB Molecule:gaf domain of unknown function;
PDBTitle: crystal structure of a gaf domain containing protein that belongs to2 pfam duf484 family (pa5279) from pseudomonas aeruginosa at 2.43 a3 resolution
|
| 54 | c3g36D_ |
|
not modelled |
85.0 |
29 |
PDB header:nuclear protein
Chain: D: PDB Molecule:protein dpy-30 homolog;
PDBTitle: crystal structure of the human dpy-30-like c-terminal domain
|
| 55 | c4e04B_ |
|
not modelled |
84.1 |
17 |
PDB header:signaling protein
Chain: B: PDB Molecule:bacteriophytochrome (light-regulated signal transduction
PDBTitle: rpbphp2 chromophore-binding domain crystallized by homologue-directed2 mutagenesis.
|
| 56 | c2au3A_ |
|
not modelled |
83.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dna primase;
PDBTitle: crystal structure of the aquifex aeolicus primase (zinc binding and2 rna polymerase domains)
|
| 57 | c2o9bA_ |
|
not modelled |
81.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:bacteriophytochrome;
PDBTitle: crystal structure of bacteriophytochrome chromophore binding domain
|
| 58 | c2xssB_ |
|
not modelled |
78.8 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:cgmp-specific 3', 5'-cyclic phosphodiesterase;
PDBTitle: crystal structure of gafb from the human phosphodiesterase 5
|
| 59 | c3su8X_ |
|
not modelled |
76.3 |
29 |
PDB header:apoptosis/signaling protein
Chain: X: PDB Molecule:plexin-b1;
PDBTitle: crystal structure of a truncated intracellular domain of plexin-b1 in2 complex with rac1
|
| 60 | c2rbgB_ |
|
not modelled |
75.3 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein st0493;
PDBTitle: crystal structure of hypothetical protein(st0493) from2 sulfolobus tokodaii
|
| 61 | c2w1tB_ |
|
not modelled |
74.8 |
17 |
PDB header:transcription
Chain: B: PDB Molecule:stage v sporulation protein t;
PDBTitle: crystal structure of b. subtilis spovt
|
| 62 | c3hm6X_ |
|
not modelled |
74.1 |
29 |
PDB header:signaling protein
Chain: X: PDB Molecule:plexin-b1;
PDBTitle: crystal structure of the cytoplasmic domain of human plexin b1
|
| 63 | c2g7uB_ |
|
not modelled |
73.9 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: 2.3 a structure of putative catechol degradative operon regulator from2 rhodococcus sp. rha1
|
| 64 | d2o9aa1 |
|
not modelled |
72.5 |
15 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:IclR ligand-binding domain-like |
| 65 | c2o0yB_ |
|
not modelled |
72.5 |
13 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator rha1_ro069532 (iclr-family) from rhodococcus sp.
|
| 66 | c3ig3A_ |
|
not modelled |
66.4 |
16 |
PDB header:signaling protein, membrane protein
Chain: A: PDB Molecule:plxna3 protein;
PDBTitle: crystal strucure of mouse plexin a3 intracellular domain
|
| 67 | d3by8a1 |
|
not modelled |
65.8 |
13 |
Fold:Profilin-like
Superfamily:Sensory domain-like
Family:Sensory domain of two-component sensor kinase |
| 68 | c2veaA_ |
|
not modelled |
63.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:phytochrome-like protein cph1;
PDBTitle: the complete sensory module of the cyanobacterial2 phytochrome cph1 in the pr-state.
|
| 69 | c2ia2D_ |
|
not modelled |
63.8 |
12 |
PDB header:transcription
Chain: D: PDB Molecule:putative transcriptional regulator;
PDBTitle: the crystal structure of a putative transcriptional regulator rha061952 from rhodococcus sp. rha1
|
| 70 | c2w1rA_ |
|
not modelled |
60.8 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:stage v sporulation protein t;
PDBTitle: crystal structure of the c-terminal domain of b. subtilis2 spovt
|
| 71 | c1mkmA_ |
|
not modelled |
60.7 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:iclr transcriptional regulator;
PDBTitle: crystal structure of the thermotoga maritima iclr
|
| 72 | c1yspA_ |
|
not modelled |
57.3 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator kdgr;
PDBTitle: crystal structure of the c-terminal domain of e. coli transcriptional2 regulator kdgr.
|
| 73 | d1tf1a_ |
|
not modelled |
56.3 |
14 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:IclR ligand-binding domain-like |
| 74 | c3hpbA_
|
|
 |
54.8 |
29 |
PDB header:protein transport
Chain: A: PDB Molecule:snx5 protein;
PDBTitle: crystal structure of snx5-px domain in p212121 space group
|
| 75 | d1guqa2 |
|
not modelled |
54.7 |
9 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 76 | d1z2wa1 |
|
not modelled |
53.5 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 77 | c3zxuC_ |
|
not modelled |
52.1 |
21 |
PDB header:cell cycle
Chain: C: PDB Molecule:mcm21;
PDBTitle: crystal structure of the ctf19-mcm21 kinetochore heterodimer from2 yeast
|
| 78 | c3fldA_ |
|
not modelled |
51.3 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein trai;
PDBTitle: crystal structure of the trai c-terminal domain
|
| 79 | c3r4kD_ |
|
not modelled |
49.6 |
15 |
PDB header:dna binding protein
Chain: D: PDB Molecule:transcriptional regulator, iclr family;
PDBTitle: crystal structure of a putative iclr transcriptional regulator2 (tm1040_3717) from silicibacter sp. tm1040 at 2.46 a resolution
|
| 80 | c3obfA_ |
|
not modelled |
49.0 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:putative transcriptional regulator, iclr family;
PDBTitle: crystal structure of putative transcriptional regulator, iclr family;2 targeted domain 129...302
|
| 81 | c3nybA_ |
|
not modelled |
46.6 |
17 |
PDB header:transferase/rna binding protein
Chain: A: PDB Molecule:poly(a) rna polymerase protein 2;
PDBTitle: structure and function of the polymerase core of tramp, a rna2 surveillance complex
|
| 82 | c3uksB_ |
|
not modelled |
43.1 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:sedoheptulose-1,7 bisphosphatase, putative;
PDBTitle: 1.85 angstrom crystal structure of putative sedoheptulose-1,72 bisphosphatase from toxoplasma gondii
|
| 83 | d1rzya_ |
|
not modelled |
43.0 |
12 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 84 | d1mkma2 |
|
not modelled |
42.2 |
12 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:IclR ligand-binding domain-like |
| 85 | c3c2wB_ |
|
not modelled |
40.4 |
9 |
PDB header:signaling protein
Chain: B: PDB Molecule:bacteriophytochrome;
PDBTitle: crystal structure of the photosensory core domain of p. aeruginosa2 bacteriophytochrome pabphp in the pfr state
|
| 86 | c3fosA_ |
|
not modelled |
40.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of two-component sensor histidine kinase domain from2 bacillus subtilis subsp. subtilis str. 168
|
| 87 | c2rajA_ |
|
not modelled |
38.9 |
37 |
PDB header:structural protein
Chain: A: PDB Molecule:sorting nexin-9;
PDBTitle: so4 bound px-bar membrane remodeling unit of sorting nexin 9
|
| 88 | d1q46a1 |
|
not modelled |
36.5 |
14 |
Fold:SAM domain-like
Superfamily:eIF2alpha middle domain-like
Family:eIF2alpha middle domain-like |
| 89 | c2hq8B_ |
|
not modelled |
36.4 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:coelenterazine-binding protein ca-bound apo form;
PDBTitle: crystal structure of coelenterazine-binding protein from2 renilla muelleri in the ca loaded apo form
|
| 90 | c4f11A_ |
|
not modelled |
35.2 |
31 |
PDB header:signaling protein
Chain: A: PDB Molecule:gamma-aminobutyric acid type b receptor subunit 2;
PDBTitle: crystal structure of the extracellular domain of human gaba(b)2 receptor gbr2
|
| 91 | d1riqa1 |
|
not modelled |
34.9 |
38 |
Fold:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Superfamily:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Family:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS) |
| 92 | c4gs4A_ |
|
not modelled |
34.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-tubulin n-acetyltransferase;
PDBTitle: structure of the alpha-tubulin acetyltransferase, alpha-tat1
|
| 93 | c3fzgA_ |
|
not modelled |
34.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:16s rrna methylase;
PDBTitle: structure of the 16s rrna methylase arma
|
| 94 | c3mq0A_ |
|
not modelled |
32.0 |
21 |
PDB header:transcription repressor
Chain: A: PDB Molecule:transcriptional repressor of the blcabc operon;
PDBTitle: crystal structure of agobacterium tumefaciens repressor blcr
|
| 95 | c2pdtD_ |
|
not modelled |
29.8 |
25 |
PDB header:circadian clock protein
Chain: D: PDB Molecule:vivid pas protein vvd;
PDBTitle: 2.3 angstrom structure of phosphodiesterase treated vivid
|
| 96 | c3cqrB_ |
|
not modelled |
29.6 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:violaxanthin de-epoxidase, chloroplast;
PDBTitle: crystal structure of the lipocalin domain of violaxanthin de-epoxidase2 (vde) at ph5
|
| 97 | c3jpyA_ |
|
not modelled |
29.5 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:glutamate [nmda] receptor subunit epsilon-2;
PDBTitle: crystal structure of the zinc-bound amino terminal domain of the nmda2 receptor subunit nr2b
|
| 98 | d1j75a_ |
|
not modelled |
29.4 |
33 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Z-DNA binding domain |
| 99 | c2gl7C_ |
|
not modelled |
29.2 |
29 |
PDB header:transcription
Chain: C: PDB Molecule:b-cell lymphoma 9 protein;
PDBTitle: crystal structure of a beta-catenin/bcl9/tcf4 complex
|
| 100 | c3sl9D_ |
|
not modelled |
29.2 |
29 |
PDB header:signaling protein, protein binding
Chain: D: PDB Molecule:b-cell cll/lymphoma 9 protein;
PDBTitle: x-ray structure of beta catenin in complex with bcl9
|
| 101 | c1ydmC_ |
|
not modelled |
28.9 |
14 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein yqgn;
PDBTitle: x-ray structure of northeast structural genomics target sr44
|
| 102 | d1y23a_ |
|
not modelled |
28.8 |
16 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 103 | c3u0kA_ |
|
not modelled |
28.6 |
14 |
PDB header:fluorescent protein
Chain: A: PDB Molecule:rcamp;
PDBTitle: crystal structure of the genetically encoded calcium indicator rcamp
|
| 104 | d2ahua2 |
|
not modelled |
28.6 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 105 | c1ysqA_ |
|
not modelled |
28.4 |
19 |
PDB header:gene regulation
Chain: A: PDB Molecule:hth-type transcriptional regulator yiaj;
PDBTitle: the crystal structure of transcriptional regulator yaij
|
| 106 | c2lnbA_ |
|
not modelled |
28.2 |
33 |
PDB header:immune system
Chain: A: PDB Molecule:z-dna-binding protein 1;
PDBTitle: solution nmr structure of n-terminal domain (6-74) of human zbp12 protein, northeast structural genomics consortium target hr8174a.
|
| 107 | d1r5la2 |
|
not modelled |
27.8 |
11 |
Fold:SpoIIaa-like
Superfamily:CRAL/TRIO domain
Family:CRAL/TRIO domain |
| 108 | d1v65a_ |
|
not modelled |
27.4 |
23 |
Fold:KRAB domain (Kruppel-associated box)
Superfamily:KRAB domain (Kruppel-associated box)
Family:KRAB domain (Kruppel-associated box) |
| 109 | d1ewqa3 |
|
not modelled |
27.3 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:DNA repair protein MutS, domain II
Family:DNA repair protein MutS, domain II |
| 110 | c2qkpD_ |
|
not modelled |
27.1 |
14 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of c-terminal domain of smu_1151c from streptococcus2 mutans
|
| 111 | c3li6G_ |
|
not modelled |
27.1 |
19 |
PDB header:metal binding protein
Chain: G: PDB Molecule:calcium-binding protein;
PDBTitle: crystal structure and trimer-monomer transition of n-terminal domain2 of ehcabp1 from entamoeba histolytica
|
| 112 | c3li6A_ |
|
not modelled |
27.1 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calcium-binding protein;
PDBTitle: crystal structure and trimer-monomer transition of n-terminal domain2 of ehcabp1 from entamoeba histolytica
|
| 113 | c3rkxA_ |
|
not modelled |
26.2 |
40 |
PDB header:ligase
Chain: A: PDB Molecule:biotin-[acetyl-coa-carboxylase] ligase;
PDBTitle: structural characterisation of staphylococcus aureus biotin protein2 ligase
|
| 114 | c2c37L_ |
|
not modelled |
26.1 |
7 |
PDB header:hydrolase
Chain: L: PDB Molecule:probable exosome complex exonuclease 1;
PDBTitle: rnase ph core of the archaeal exosome in complex with u82 rna
|
| 115 | d1g33a_ |
|
not modelled |
25.6 |
24 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Parvalbumin |
| 116 | c1ih0A_ |
|
not modelled |
25.2 |
24 |
PDB header:contractile protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: structure of the c-domain of human cardiac troponin c in2 complex with ca2+ sensitizer emd 57033
|
| 117 | d1ih0a_ |
|
not modelled |
25.2 |
24 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 118 | c2aexA_ |
|
not modelled |
25.1 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coproporphyrinogen iii oxidase, mitochondrial;
PDBTitle: the 1.58a crystal structure of human coproporphyrinogen oxidase2 reveals the structural basis of hereditary coproporphyria
|
| 119 | c4ienB_ |
|
not modelled |
25.0 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative acyl-coa hydrolase;
PDBTitle: crystal structure of acyl-coa hydrolase from neisseria meningitidis2 fam18
|
| 120 | d1txna_ |
|
not modelled |
24.9 |
25 |
Fold:Coproporphyrinogen III oxidase
Superfamily:Coproporphyrinogen III oxidase
Family:Coproporphyrinogen III oxidase |