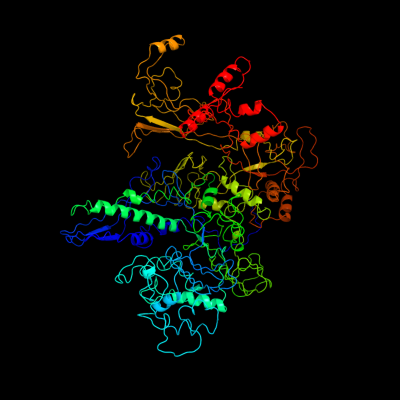
| 1 | c3iydC_
|
|
 |
100.0 |
99 |
PDB header:transcription/dna
Chain: C: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
|
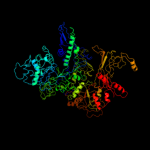
| 2 | d1smyc_
|
|
 |
100.0 |
50 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
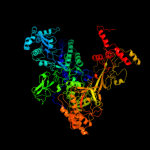
| 3 | d1ynjc1
|
|
 |
100.0 |
50 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
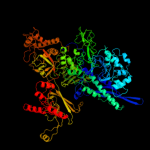
| 4 | c2pmzB_
|
|
 |
100.0 |
31 |
PDB header:translation, transferase
Chain: B: PDB Molecule:dna-directed rna polymerase subunit b;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 5 | c3h0gN_
|
|
 |
100.0 |
27 |
PDB header:transcription
Chain: N: PDB Molecule:dna-directed rna polymerase ii subunit rpb2;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 6 | d1twfb_
|
|
 |
100.0 |
30 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
| 7 | c3mlqD_
|
|
 |
100.0 |
54 |
PDB header:transferase/transcription
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
|
| 8 | c3mlqB_
|
|
 |
100.0 |
59 |
PDB header:transferase/transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
|
| 9 | c3tbiB_
|
|
 |
100.0 |
100 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of t4 gp33 bound to e. coli rnap beta-flap domain
|
| 10 | c3ltiA_
|
|
 |
100.0 |
92 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of the escherichia coli rna polymerase beta subunit2 beta2-betai4 domains
|
| 11 | c3qqcA_
|
|
 |
99.4 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit b, dna-directed rna
PDBTitle: crystal structure of archaeal spt4/5 bound to the rnap clamp domain
|
| 12 | c3e7hA_
|
|
 |
95.6 |
75 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: the crystal structure of the beta subunit of the dna-2 directed rna polymerase from vibrio cholerae o1 biovar3 eltor
|
| 13 | d1r46a1
|
|
 |
50.9 |
20 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 14 | c1gjjA_
|
|
 |
50.3 |
20 |
PDB header:membrane protein
Chain: A: PDB Molecule:lap2;
PDBTitle: n-terminal constant region of the nuclear envelope protein2 lap2
|
| 15 | c2gtiA_
|
|
 |
41.6 |
14 |
PDB header:viral protein
Chain: A: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: mutation of mhv coronavirus non-structural protein nsp15 (f307l)
|
| 16 | d1n26a2
|
|
 |
30.8 |
11 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
| 17 | d2eyqa5
|
|
 |
28.1 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 18 | d1gm5a4
|
|
 |
27.8 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 19 | d1y4oa1
|
|
 |
26.6 |
53 |
Fold:Profilin-like
Superfamily:Roadblock/LC7 domain
Family:Roadblock/LC7 domain |
| 20 | d2f3na1
|
|
 |
25.7 |
10 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 21 | d1zq1c1 |
|
not modelled |
24.5 |
36 |
Fold:GatB/YqeY motif
Superfamily:GatB/YqeY motif
Family:GatB/GatE C-terminal domain-like |
| 22 | c2db3D_ |
|
not modelled |
24.0 |
38 |
PDB header:hydrolase/rna
Chain: D: PDB Molecule:atp-dependent rna helicase vasa;
PDBTitle: structural basis for rna unwinding by the dead-box protein2 drosophila vasa
|
| 23 | d2g7ja1 |
|
not modelled |
22.8 |
32 |
Fold:Secretion chaperone-like
Superfamily:YgaC/TfoX-N like
Family:YgaC-like |
| 24 | d1nbwa1 |
|
not modelled |
21.4 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Swiveling domain of dehydratase reactivase alpha subunit
Family:Swiveling domain of dehydratase reactivase alpha subunit |
| 25 | d2gtia2 |
|
not modelled |
21.2 |
10 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Nsp15 C-terminal domain-like |
| 26 | c2c2lD_ |
|
not modelled |
20.5 |
19 |
PDB header:chaperone
Chain: D: PDB Molecule:carboxy terminus of hsp70-interacting protein;
PDBTitle: crystal structure of the chip u-box e3 ubiquitin ligase
|
| 27 | d1sy7a1 |
|
not modelled |
20.5 |
15 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Catalase, C-terminal domain |
| 28 | c2aujD_ |
|
not modelled |
20.4 |
33 |
PDB header:transferase
Chain: D: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of thermus aquaticus rna polymerase beta'-subunit2 insert
|
| 29 | c3mjhD_ |
|
not modelled |
19.3 |
38 |
PDB header:protein transport
Chain: D: PDB Molecule:early endosome antigen 1;
PDBTitle: crystal structure of human rab5a in complex with the c2h2 zinc finger2 of eea1
|
| 30 | c3bxzA_ |
|
not modelled |
19.2 |
33 |
PDB header:transport protein
Chain: A: PDB Molecule:preprotein translocase subunit seca;
PDBTitle: crystal structure of the isolated dead motor domains from2 escherichia coli seca
|
| 31 | c3cwiA_ |
|
not modelled |
18.9 |
22 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:thiamine-biosynthesis protein this;
PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
|
| 32 | d1d02a_ |
|
not modelled |
18.7 |
17 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Restriction endonuclease MunI |
| 33 | d1ujpa_ |
|
not modelled |
18.4 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 34 | c3izbX_ |
|
not modelled |
18.3 |
63 |
PDB header:ribosome
Chain: X: PDB Molecule:40s ribosomal protein rps27 (s27e);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 35 | d1jqna_ |
|
not modelled |
18.1 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate carboxylase |
| 36 | d1tf5a4 |
|
not modelled |
17.7 |
33 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 37 | c3kdeC_ |
|
not modelled |
17.6 |
19 |
PDB header:dna binding protein/dna
Chain: C: PDB Molecule:transposable element p transposase;
PDBTitle: crystal structure of the thap domain from d. melanogaster p-element2 transposase in complex with its natural dna binding site
|
| 38 | c2nu9E_ |
|
not modelled |
16.8 |
18 |
PDB header:ligase
Chain: E: PDB Molecule:succinyl-coa synthetase beta chain;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase2 orthorhombic crystal form
|
| 39 | c3bq7A_ |
|
not modelled |
16.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:diacylglycerol kinase delta;
PDBTitle: sam domain of diacylglycerol kinase delta1 (e35g)
|
| 40 | c2hsiB_ |
|
not modelled |
16.2 |
25 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative peptidase m23;
PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
|
| 41 | d1cf2o2 |
|
not modelled |
16.0 |
31 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 42 | c3lvwA_ |
|
not modelled |
16.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:glutamate--cysteine ligase;
PDBTitle: glutathione-inhibited scgcl
|
| 43 | c3u5cI_ |
|
not modelled |
15.4 |
22 |
PDB header:ribosome
Chain: I: PDB Molecule:40s ribosomal protein s8-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
|
| 44 | c2ivfC_ |
|
not modelled |
15.3 |
38 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:ethylbenzene dehydrogenase gamma-subunit;
PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
|
| 45 | c2bhmE_ |
|
not modelled |
15.2 |
24 |
PDB header:bacterial protein
Chain: E: PDB Molecule:type iv secretion system protein virb8;
PDBTitle: crystal structure of virb8 from brucella suis
|
| 46 | c3iz6X_ |
|
not modelled |
15.2 |
75 |
PDB header:ribosome
Chain: X: PDB Molecule:40s ribosomal protein s27 (s27e);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 47 | c6paxA_ |
|
not modelled |
14.3 |
22 |
PDB header:gene regulation/dna
Chain: A: PDB Molecule:homeobox protein pax-6;
PDBTitle: crystal structure of the human pax-6 paired domain-dna2 complex reveals a general model for pax protein-dna3 interactions
|
| 48 | d2bpa2_ |
|
not modelled |
14.1 |
31 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:ssDNA viruses
Family:Microviridae-like VP |
| 49 | d2d6fc2 |
|
not modelled |
13.6 |
46 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
| 50 | c2vb0A_ |
|
not modelled |
13.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:polyprotein 3bcd;
PDBTitle: crystal structure of coxsackievirus b3 proteinase 3c
|
| 51 | d2hz5a1 |
|
not modelled |
13.1 |
59 |
Fold:Profilin-like
Superfamily:Roadblock/LC7 domain
Family:Roadblock/LC7 domain |
| 52 | c3eq5G_ |
|
not modelled |
12.9 |
18 |
PDB header:signaling protein
Chain: G: PDB Molecule:ski-like protein;
PDBTitle: crystal structure of fragment 137 to 238 of the human ski-like protein
|
| 53 | c1m1jA_ |
|
not modelled |
12.9 |
38 |
PDB header:blood clotting
Chain: A: PDB Molecule:fibrinogen alpha subunit;
PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
|
| 54 | d2gpra_ |
|
not modelled |
12.6 |
17 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
| 55 | d1dcza_ |
|
not modelled |
12.4 |
27 |
Fold:Barrel-sandwich hybrid
Superfamily:Single hybrid motif
Family:Biotinyl/lipoyl-carrier proteins and domains |
| 56 | d1flja_ |
|
not modelled |
12.2 |
33 |
Fold:Carbonic anhydrase
Superfamily:Carbonic anhydrase
Family:Carbonic anhydrase |
| 57 | d2pxyd2 |
|
not modelled |
12.2 |
56 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 58 | c3dlmA_ |
|
not modelled |
12.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:histone-lysine n-methyltransferase setdb1;
PDBTitle: crystal structure of tudor domain of human histone-lysine n-2 methyltransferase setdb1
|
| 59 | c2qm3A_ |
|
not modelled |
11.8 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of a predicted methyltransferase from pyrococcus2 furiosus
|
| 60 | d1px5a1 |
|
not modelled |
11.7 |
28 |
Fold:PAP/OAS1 substrate-binding domain
Superfamily:PAP/OAS1 substrate-binding domain
Family:2'-5'-oligoadenylate synthetase 1, OAS1, second domain |
| 61 | c2cwpA_ |
|
not modelled |
11.7 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:metrs related protein;
PDBTitle: crystal structure of metrs related protein from pyrococcus horikoshii
|
| 62 | c2gu1A_ |
|
not modelled |
11.4 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:zinc peptidase;
PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
|
| 63 | d1lk5a2 |
|
not modelled |
11.4 |
19 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 64 | d1pfva2 |
|
not modelled |
11.3 |
40 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
| 65 | d2pwwa1 |
|
not modelled |
11.2 |
7 |
Fold:TBP-like
Superfamily:YugN-like
Family:YugN-like |
| 66 | d1prtc1 |
|
not modelled |
11.1 |
14 |
Fold:OB-fold
Superfamily:Bacterial enterotoxins
Family:Bacterial AB5 toxins, B-subunits |
| 67 | c2f1mA_ |
|
not modelled |
11.1 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:acriflavine resistance protein a;
PDBTitle: conformational flexibility in the multidrug efflux system protein acra
|
| 68 | d1glaf_ |
|
not modelled |
10.8 |
20 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
| 69 | c2oy7A_ |
|
not modelled |
10.7 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:outer surface protein a;
PDBTitle: the crystal structure of ospa mutant
|
| 70 | d1gjja1 |
|
not modelled |
10.5 |
29 |
Fold:LEM/SAP HeH motif
Superfamily:LEM domain
Family:LEM domain |
| 71 | c2hxyC_ |
|
not modelled |
10.3 |
33 |
PDB header:hydrolase
Chain: C: PDB Molecule:probable atp-dependent rna helicase ddx48;
PDBTitle: crystal structure of human apo-eif4aiii
|
| 72 | c2dneA_ |
|
not modelled |
10.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dihydrolipoyllysine-residue acetyltransferase
PDBTitle: solution structure of rsgi ruh-058, a lipoyl domain of2 human 2-oxoacid dehydrogenase
|
| 73 | c2pjmA_ |
|
not modelled |
10.2 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
|
| 74 | c3nyyA_ |
|
not modelled |
10.2 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative glycyl-glycine endopeptidase lytm;
PDBTitle: crystal structure of a putative glycyl-glycine endopeptidase lytm2 (rumgna_02482) from ruminococcus gnavus atcc 29149 at 1.60 a3 resolution
|
| 75 | d1m0sa2 |
|
not modelled |
10.2 |
19 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 76 | d1pk1c1 |
|
not modelled |
10.2 |
3 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 77 | d1f46a_ |
|
not modelled |
10.1 |
57 |
Fold:TBP-like
Superfamily:Cell-division protein ZipA, C-terminal domain
Family:Cell-division protein ZipA, C-terminal domain |
| 78 | c3ghgD_ |
|
not modelled |
10.0 |
29 |
PDB header:blood clotting
Chain: D: PDB Molecule:fibrinogen alpha chain;
PDBTitle: crystal structure of human fibrinogen
|
| 79 | c1pk1A_ |
|
not modelled |
9.9 |
5 |
PDB header:transcription repression
Chain: A: PDB Molecule:polyhomeotic-proximal chromatin protein;
PDBTitle: hetero sam domain structure of ph and scm.
|
| 80 | c1pd7B_ |
|
not modelled |
9.9 |
60 |
PDB header:transcription
Chain: B: PDB Molecule:mad1;
PDBTitle: extended sid of mad1 bound to the pah2 domain of msin3b
|
| 81 | d1aola_ |
|
not modelled |
9.8 |
21 |
Fold:ENV polyprotein, receptor-binding domain
Superfamily:ENV polyprotein, receptor-binding domain
Family:ENV polyprotein, receptor-binding domain |
| 82 | c2jgnB_ |
|
not modelled |
9.7 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:atp-dependent rna helicase ddx3x;
PDBTitle: ddx3 helicase domain
|
| 83 | c3gekA_ |
|
not modelled |
9.7 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative thioesterase yhda;
PDBTitle: crystal structure of putative thioesterase yhda from lactococcus2 lactis. northeast structural genomics consortium target kr113
|
| 84 | d1prtb1 |
|
not modelled |
9.7 |
14 |
Fold:OB-fold
Superfamily:Bacterial enterotoxins
Family:Bacterial AB5 toxins, B-subunits |
| 85 | d1luga_ |
|
not modelled |
9.6 |
33 |
Fold:Carbonic anhydrase
Superfamily:Carbonic anhydrase
Family:Carbonic anhydrase |
| 86 | d2c4ka1 |
|
not modelled |
9.5 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 87 | c3kizA_ |
|
not modelled |
9.5 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of putative phosphoribosylformylglycinamidine cyclo-2 ligase (yp_676759.1) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
|
| 88 | c1a1vA_ |
|
not modelled |
9.5 |
26 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:protein (ns3 protein);
PDBTitle: hepatitis c virus ns3 helicase domain complexed with single2 stranded sdna
|
| 89 | d1m5ha1 |
|
not modelled |
9.5 |
42 |
Fold:Ferredoxin-like
Superfamily:Formylmethanofuran:tetrahydromethanopterin formyltransferase
Family:Formylmethanofuran:tetrahydromethanopterin formyltransferase |
| 90 | d2d0oa1 |
|
not modelled |
9.4 |
12 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Swiveling domain of dehydratase reactivase alpha subunit
Family:Swiveling domain of dehydratase reactivase alpha subunit |
| 91 | d1u9ya1 |
|
not modelled |
9.4 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 92 | d1ftra1 |
|
not modelled |
9.4 |
54 |
Fold:Ferredoxin-like
Superfamily:Formylmethanofuran:tetrahydromethanopterin formyltransferase
Family:Formylmethanofuran:tetrahydromethanopterin formyltransferase |
| 93 | d1gpra_ |
|
not modelled |
9.3 |
25 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
| 94 | d1vf7a_ |
|
not modelled |
9.3 |
27 |
Fold:HlyD-like secretion proteins
Superfamily:HlyD-like secretion proteins
Family:HlyD-like secretion proteins |
| 95 | c2xzm6_ |
|
not modelled |
9.2 |
63 |
PDB header:ribosome
Chain: 6: PDB Molecule:rps27e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 96 | d1m5sa1 |
|
not modelled |
9.2 |
42 |
Fold:Ferredoxin-like
Superfamily:Formylmethanofuran:tetrahydromethanopterin formyltransferase
Family:Formylmethanofuran:tetrahydromethanopterin formyltransferase |
| 97 | d1qxfa_ |
|
not modelled |
9.2 |
33 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein S27e |
| 98 | c2kkpA_ |
|
not modelled |
9.2 |
36 |
PDB header:dna binding protein
Chain: A: PDB Molecule:phage integrase;
PDBTitle: solution nmr structure of the phage integrase sam-like2 domain from moth 1796 from moorella thermoacetica.3 northeast structural genomics consortium target mtr39k4 (residues 64-171).
|
| 99 | c2fyuE_ |
|
not modelled |
9.1 |
43 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|