| 1 | c2qgqF_
|
|
 |
100.0 |
32 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:protein tm_1862;
PDBTitle: crystal structure of tm_1862 from thermotoga maritima.2 northeast structural genomics consortium target vr77
|
| 2 | c3cixA_
|
|
 |
100.0 |
15 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
| 3 | d1olta_
|
|
 |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
| 4 | c3t7vA_
|
|
 |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
| 5 | c1r30A_
|
|
 |
99.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
|
| 6 | d1r30a_
|
|
 |
99.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
| 7 | c3rfaA_
|
|
 |
99.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
| 8 | c2yx0A_
|
|
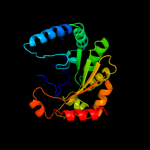 |
99.7 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
| 9 | d1tv8a_
|
|
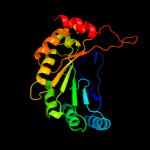 |
99.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:MoCo biosynthesis proteins |
| 10 | c2a5hC_
|
|
 |
99.5 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
| 11 | c3c8fA_
|
|
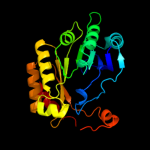 |
99.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate formate-lyase 1-activating enzyme;
PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
|
| 12 | c2z2uA_
|
|
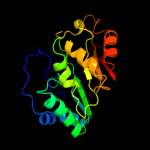 |
97.4 |
10 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
| 13 | c3canA_
|
|
 |
97.1 |
14 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
| 14 | c2ftpA_
|
|
 |
96.4 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
| 15 | d3bula2
|
|
 |
95.3 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 16 | d7reqa2
|
|
 |
95.2 |
19 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 17 | d1yvca1
|
|
 |
95.0 |
18 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:TRAM domain |
| 18 | d1yeza1
|
|
 |
95.0 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:TRAM domain |
| 19 | c1bmtB_
|
|
 |
94.9 |
16 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
| 20 | c2cw6B_
|
|
 |
94.8 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
| 21 | d1uwva1 |
|
not modelled |
94.4 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:TRAM domain |
| 22 | c1uwvA_ |
|
not modelled |
94.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:23s rrna (uracil-5-)-methyltransferase ruma;
PDBTitle: crystal structure of ruma, the iron-sulfur cluster2 containing e. coli 23s ribosomal rna 5-methyluridine3 methyltransferase
|
| 23 | c3ivuB_ |
|
not modelled |
94.3 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 24 | c1y80A_
|
|
 |
93.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 25 | d1f6ya_ |
|
not modelled |
93.9 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 26 | c3eegB_ |
|
not modelled |
93.2 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 27 | c1ydnA_ |
|
not modelled |
93.0 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 28 | c3ewbX_ |
|
not modelled |
92.8 |
10 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
|
| 29 | d1qt1a_ |
|
not modelled |
92.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 30 | c1e1cA_
|
|
 |
92.5 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
| 31 | d1nvma2 |
|
not modelled |
92.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 32 | d1ccwa_
|
|
 |
92.1 |
14 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 33 | c1nvmG_ |
|
not modelled |
91.5 |
13 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 34 | c1tx2A_ |
|
not modelled |
90.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 35 | d1tx2a_ |
|
not modelled |
90.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 36 | c2vs1A_ |
|
not modelled |
90.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized rna methyltransferase pyrab10780;
PDBTitle: the crystal structure of pyrococcus abyssi trna (uracil-54,2 c5)-methyltransferase in complex with s-adenosyl-l-3 homocysteine
|
| 37 | c2p10D_ |
|
not modelled |
90.8 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 38 | d1muwa_ |
|
not modelled |
90.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 39 | c3ezxA_ |
|
not modelled |
90.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
| 40 | c2yciX_ |
|
not modelled |
88.9 |
15 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 41 | c3khdC_ |
|
not modelled |
88.4 |
12 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pff1300w.
|
| 42 | d1fmfa_ |
|
not modelled |
88.4 |
18 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 43 | d1xima_ |
|
not modelled |
87.8 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 44 | c3bleA_ |
|
not modelled |
85.6 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 45 | c3hpxB_ |
|
not modelled |
85.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of mycobacterium tuberculosis leua active site2 domain 1-425 (truncation mutant delta:426-644)
|
| 46 | d2glka1 |
|
not modelled |
84.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 47 | c1zfjA_ |
|
not modelled |
83.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 48 | c3tr9A_ |
|
not modelled |
83.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 49 | c2bdqA_ |
|
not modelled |
82.2 |
13 |
PDB header:metal transport
Chain: A: PDB Molecule:copper homeostasis protein cutc;
PDBTitle: crystal structure of the putative copper homeostasis2 protein cutc from streptococcus agalactiae, northeast3 strucural genomics target sar15.
|
| 50 | c2i2xD_ |
|
not modelled |
81.6 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
| 51 | c2yxbA_ |
|
not modelled |
81.6 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 52 | c1k98A_ |
|
not modelled |
81.6 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
| 53 | d1ad1a_ |
|
not modelled |
81.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 54 | c2hk1D_ |
|
not modelled |
81.0 |
12 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
| 55 | d1h4pa_ |
|
not modelled |
80.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 56 | d1eyea_ |
|
not modelled |
79.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 57 | c3ct7E_ |
|
not modelled |
77.4 |
14 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
| 58 | d1mxsa_ |
|
not modelled |
76.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 59 | d3bofa1 |
|
not modelled |
76.5 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 60 | d1xrsb1 |
|
not modelled |
76.1 |
20 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 61 | d1k77a_ |
|
not modelled |
76.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Hypothetical protein YgbM (EC1530) |
| 62 | c2yw3E_ |
|
not modelled |
76.0 |
18 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
| 63 | d1vhca_ |
|
not modelled |
74.6 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 64 | c3no5C_ |
|
not modelled |
74.3 |
12 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
|
| 65 | d1pkla2 |
|
not modelled |
74.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 66 | c3lu2B_ |
|
not modelled |
74.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2462 protein;
PDBTitle: structure of lmo2462, a listeria monocytogenes amidohydrolase family2 putative dipeptidase
|
| 67 | d1ajza_ |
|
not modelled |
73.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 68 | c3bicA_ |
|
not modelled |
73.6 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase, mitochondrial precursor;
PDBTitle: crystal structure of human methylmalonyl-coa mutase
|
| 69 | d2fiqa1 |
|
not modelled |
72.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:GatZ-like |
| 70 | c1sr9A_ |
|
not modelled |
71.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of leua from mycobacterium tuberculosis
|
| 71 | d1xlma_ |
|
not modelled |
70.8 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 72 | c6reqB_ |
|
not modelled |
70.6 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:protein (methylmalonyl-coa mutase);
PDBTitle: methylmalonyl-coa mutase, 3-carboxypropyl-coa inhibitor complex
|
| 73 | d1x7fa2 |
|
not modelled |
69.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Outer surface protein, N-terminal domain |
| 74 | c3npgD_ |
|
not modelled |
69.2 |
15 |
PDB header:unknown function
Chain: D: PDB Molecule:uncharacterized duf364 family protein;
PDBTitle: crystal structure of a protein with unknown function from duf3642 family (ph1506) from pyrococcus horikoshii at 2.70 a resolution
|
| 75 | c3bolB_ |
|
not modelled |
68.8 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 76 | d2h1qa1 |
|
not modelled |
68.4 |
21 |
Fold:PLP-dependent transferase-like
Superfamily:Dhaf3308-like
Family:Dhaf3308-like |
| 77 | c3itcA_ |
|
not modelled |
68.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:renal dipeptidase;
PDBTitle: crystal structure of sco3058 with bound citrate and glycerol
|
| 78 | d1tqxa_ |
|
not modelled |
66.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 79 | c3r79B_ |
|
not modelled |
66.4 |
11 |
PDB header:structure genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharactertized protein from agrobacterium2 tumefaciens
|
| 80 | c3labA_ |
|
not modelled |
65.6 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate)
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
| 81 | c1ydoC_ |
|
not modelled |
65.3 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
| 82 | d1qopa_ |
|
not modelled |
65.3 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 83 | d1wbha1 |
|
not modelled |
65.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 84 | d1itua_ |
|
not modelled |
64.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Renal dipeptidase |
| 85 | c3fdgA_ |
|
not modelled |
63.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidase ac. metallo peptidase. merops family m19;
PDBTitle: the crystal structure of the dipeptidase ac, metallo peptidase. merops2 family m19
|
| 86 | c1x7fA_ |
|
not modelled |
63.1 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:outer surface protein;
PDBTitle: crystal structure of an uncharacterized b. cereus protein
|
| 87 | d1liua2 |
|
not modelled |
62.8 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 88 | d1e0ta2 |
|
not modelled |
62.6 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 89 | d1mvoa_ |
|
not modelled |
62.5 |
17 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 90 | d1bxca_ |
|
not modelled |
61.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 91 | c2vefB_ |
|
not modelled |
60.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 92 | c3ngfA_ |
|
not modelled |
60.4 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family 2;
PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
|
| 93 | c2qs0A_ |
|
not modelled |
60.2 |
20 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:quinolinate synthetase a;
PDBTitle: quinolinate synthase from pyrococcus furiosus
|
| 94 | c1u5tA_ |
|
not modelled |
60.0 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:appears to be functionally related to snf7;
PDBTitle: structure of the escrt-ii endosomal trafficking complex
|
| 95 | d1gvia3 |
|
not modelled |
59.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 96 | c2zyfA_ |
|
not modelled |
59.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:homocitrate synthase;
PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with magnesuim ion and alpha-ketoglutarate
|
| 97 | c2xvzA_ |
|
not modelled |
56.6 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:chelatase, putative;
PDBTitle: cobalt chelatase cbik (periplasmatic) from desulvobrio2 vulgaris hildenborough (co-crystallized with cobalt)
|
| 98 | c3cqkB_ |
|
not modelled |
56.1 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 99 | c3chvA_ |
|
not modelled |
55.3 |
11 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
| 100 | d1mkza_ |
|
not modelled |
55.2 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 101 | c2vp8A_ |
|
not modelled |
55.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
| 102 | d1h1ya_ |
|
not modelled |
55.0 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 103 | c1xrsB_ |
|
not modelled |
54.6 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:d-lysine 5,6-aminomutase beta subunit;
PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
|
| 104 | d1a3xa2 |
|
not modelled |
54.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 105 | d1m7xa3 |
|
not modelled |
53.1 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 106 | d2pb1a1 |
|
not modelled |
52.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 107 | c3dx5A_ |
|
not modelled |
52.9 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 108 | c3elbA_ |
|
not modelled |
52.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:ethanolamine-phosphate cytidylyltransferase;
PDBTitle: human ctp: phosphoethanolamine cytidylyltransferase in complex with2 cmp
|
| 109 | c3zu0A_ |
|
not modelled |
50.8 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
| 110 | d7reqb2 |
|
not modelled |
50.6 |
14 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 111 | c2i5gB_ |
|
not modelled |
50.2 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:amidohydrolase;
PDBTitle: crystal strcuture of amidohydrolase from pseudomonas2 aeruginosa
|
| 112 | d1yx1a1 |
|
not modelled |
49.0 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
| 113 | c3k1dA_ |
|
not modelled |
48.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:1,4-alpha-glucan-branching enzyme;
PDBTitle: crystal structure of glycogen branching enzyme synonym: 1,4-alpha-d-2 glucan:1,4-alpha-d-glucan 6-glucosyl-transferase from mycobacterium3 tuberculosis h37rv
|
| 114 | c3pdiG_ |
|
not modelled |
48.5 |
20 |
PDB header:protein binding
Chain: G: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nife;
PDBTitle: precursor bound nifen
|
| 115 | c1l2fA_ |
|
not modelled |
48.3 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:n utilization substance protein a;
PDBTitle: crystal structure of nusa from thermotoga maritima: a2 structure-based role of the n-terminal domain
|
| 116 | c3dhuC_ |
|
not modelled |
47.9 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure of an alpha-amylase from lactobacillus2 plantarum
|
| 117 | d1u2za_ |
|
not modelled |
47.9 |
34 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Catalytic, N-terminal domain of histone methyltransferase Dot1l |
| 118 | d1o4ua1 |
|
not modelled |
47.8 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 119 | c2d4aC_ |
|
not modelled |
47.5 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of the malate dehydrogenase from aeropyrum pernix
|
| 120 | c3pdiB_ |
|
not modelled |
46.6 |
20 |
PDB header:protein binding
Chain: B: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nifn;
PDBTitle: precursor bound nifen
|