| 1 | c3ojcD_
|
|
 |
100.0 |
57 |
PDB header:isomerase
Chain: D: PDB Molecule:putative aspartate/glutamate racemase;
PDBTitle: crystal structure of a putative asp/glu racemase from yersinia pestis
|
| 2 | c2dx7B_
|
|
 |
100.0 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:aspartate racemase;
PDBTitle: crystal structure of pyrococcus horikoshii ot3 aspartate racemase2 complex with citric acid
|
| 3 | c2zskA_
|
|
 |
100.0 |
34 |
PDB header:unknown function
Chain: A: PDB Molecule:226aa long hypothetical aspartate racemase;
PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
|
| 4 | c3s81A_
|
|
 |
100.0 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
|
| 5 | c1zuwA_
|
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase 1;
PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
|
| 6 | c3hfrA_
|
|
 |
100.0 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
|
| 7 | c2gzmB_
|
|
 |
100.0 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of the glutamate racemase from bacillus2 anthracis
|
| 8 | c2dwuA_
|
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
|
| 9 | c2jfoB_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
|
| 10 | c2jfqA_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of staphylococcus aureus glutamate2 racemase in complex with d-glutamate
|
| 11 | c3outC_
|
|
 |
100.0 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
|
| 12 | c1b74A_
|
|
 |
100.0 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase from aquifex pyrophilus
|
| 13 | c2jfzB_
|
|
 |
100.0 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
|
| 14 | c2jfnA_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
|
| 15 | c2ohoA_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: structural basis for glutamate racemase inhibitor
|
| 16 | d1jfla1
|
|
 |
100.0 |
23 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 17 | d1jfla2
|
|
 |
99.9 |
25 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 18 | c2eq5D_
|
|
 |
99.9 |
19 |
PDB header:isomerase
Chain: D: PDB Molecule:228aa long hypothetical hydantoin racemase;
PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
|
| 19 | d2dx7a1
|
|
 |
99.9 |
23 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 20 | c3qvjB_
|
|
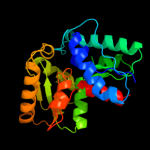 |
99.9 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
| 21 | c2vlbC_ |
|
not modelled |
99.9 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 22 | d1b74a2 |
|
not modelled |
99.8 |
18 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 23 | c2dgdD_ |
|
not modelled |
99.7 |
8 |
PDB header:lyase
Chain: D: PDB Molecule:223aa long hypothetical arylmalonate decarboxylase;
PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
|
| 24 | d1b74a1 |
|
not modelled |
99.7 |
20 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 25 | c2xecD_ |
|
not modelled |
98.8 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 26 | d1m3ua_ |
|
not modelled |
95.8 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 27 | c2q5cA_ |
|
not modelled |
95.8 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 28 | d1oy0a_ |
|
not modelled |
94.7 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 29 | c3ez4B_ |
|
not modelled |
94.5 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 30 | d1o66a_ |
|
not modelled |
94.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 31 | d2pjua1 |
|
not modelled |
94.4 |
14 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 32 | c2pjuD_ |
|
not modelled |
93.4 |
13 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 33 | c2iksA_ |
|
not modelled |
92.7 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:dna-binding transcriptional dual regulator;
PDBTitle: crystal structure of n-terminal truncated dna-binding transcriptional2 dual regulator from escherichia coli k12
|
| 34 | c1dkrB_ |
|
not modelled |
92.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyl pyrophosphate synthetase;
PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
|
| 35 | c3eooL_ |
|
not modelled |
91.7 |
15 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 36 | c3dahB_ |
|
not modelled |
91.6 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: 2.3 a crystal structure of ribose-phosphate pyrophosphokinase from2 burkholderia pseudomallei
|
| 37 | d1muma_ |
|
not modelled |
91.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 38 | d2pv7a2 |
|
not modelled |
90.9 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 39 | d1dbqa_ |
|
not modelled |
90.7 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 40 | c3i09A_ |
|
not modelled |
90.5 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic branched-chain amino acid-binding protein;
PDBTitle: crystal structure of a periplasmic binding protein (bma2936) from2 burkholderia mallei at 1.80 a resolution
|
| 41 | c2qiwA_ |
|
not modelled |
90.2 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 42 | c3ih1A_ |
|
not modelled |
89.6 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 43 | c3h5oB_ |
|
not modelled |
89.5 |
15 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator gntr;
PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
|
| 44 | c3noyA_ |
|
not modelled |
89.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
| 45 | c3sg0A_ |
|
not modelled |
88.0 |
8 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: the crystal structure of an extracellular ligand-binding receptor from2 rhodopseudomonas palustris haa2
|
| 46 | c2iswB_ |
|
not modelled |
87.3 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-1,6-bisphosphate aldolase;
PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
|
| 47 | c1u9yD_ |
|
not modelled |
87.0 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
|
| 48 | c2ze3A_ |
|
not modelled |
86.6 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 49 | d1pqua1 |
|
not modelled |
86.5 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 50 | c3d8uA_ |
|
not modelled |
86.3 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:purr transcriptional regulator;
PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
|
| 51 | c3kwlA_ |
|
not modelled |
86.1 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a hypothetical protein from helicobacter pylori
|
| 52 | c3brqA_ |
|
not modelled |
85.9 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator ascg;
PDBTitle: crystal structure of the escherichia coli transcriptional repressor2 ascg
|
| 53 | c3lftA_ |
|
not modelled |
84.7 |
15 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the abc domain in complex with l-trp from2 streptococcus pneumonia to 1.35a
|
| 54 | d1ujqa_ |
|
not modelled |
84.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 55 | c3oa2B_ |
|
not modelled |
83.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from pseudomonas2 aeruginosa in complex with nad at 1.5 angstrom resolution
|
| 56 | c2c4kD_ |
|
not modelled |
82.6 |
14 |
PDB header:regulatory protein
Chain: D: PDB Molecule:phosphoribosyl pyrophosphate synthetase-
PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
|
| 57 | c2rgyA_ |
|
not modelled |
82.5 |
15 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of transcriptional regulator of laci family from2 burkhoderia phymatum
|
| 58 | c1zlpA_ |
|
not modelled |
81.9 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 59 | d1twda_ |
|
not modelled |
81.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:CutC-like
Family:CutC-like |
| 60 | c3snrA_ |
|
not modelled |
80.2 |
8 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: rpd_1889 protein, an extracellular ligand-binding receptor from2 rhodopseudomonas palustris.
|
| 61 | c3mizB_ |
|
not modelled |
79.8 |
16 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator protein, laci
PDBTitle: crystal structure of a putative transcriptional regulator2 protein, lacl family from rhizobium etli
|
| 62 | c3orsD_ |
|
not modelled |
78.4 |
13 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
| 63 | c1xeaD_ |
|
not modelled |
78.0 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
| 64 | d1ydwa1 |
|
not modelled |
77.3 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 65 | d1o7ja_ |
|
not modelled |
76.9 |
14 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 66 | d4pgaa_ |
|
not modelled |
76.9 |
16 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 67 | c3kc2A_ |
|
not modelled |
76.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein ykr070w;
PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
|
| 68 | c3efhB_ |
|
not modelled |
75.2 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase 1;
PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
|
| 69 | c3k9cA_ |
|
not modelled |
75.1 |
15 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family protein;
PDBTitle: crystal structure of laci transcriptional regulator from rhodococcus2 species.
|
| 70 | c1i36A_ |
|
not modelled |
74.9 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein mth1747;
PDBTitle: structure of conserved protein mth1747 of unknown function2 reveals structural similarity with 3-hydroxyacid3 dehydrogenases
|
| 71 | c1ps9A_ |
|
not modelled |
74.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 72 | c3ic5A_ |
|
not modelled |
74.7 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 73 | c3hf3A_ |
|
not modelled |
74.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 74 | d1gvfa_ |
|
not modelled |
74.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 75 | c3iwpK_ |
|
not modelled |
73.5 |
33 |
PDB header:metal binding protein
Chain: K: PDB Molecule:copper homeostasis protein cutc homolog;
PDBTitle: crystal structure of human copper homeostasis protein cutc
|
| 76 | c3b8iF_ |
|
not modelled |
73.3 |
18 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 77 | c3h5lB_ |
|
not modelled |
72.4 |
12 |
PDB header:transport protein
Chain: B: PDB Molecule:putative branched-chain amino acid abc
PDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from silicibacter pomeroyi
|
| 78 | c3zu0A_ |
|
not modelled |
70.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
| 79 | c2pv7B_ |
|
not modelled |
70.8 |
13 |
PDB header:isomerase, oxidoreductase
Chain: B: PDB Molecule:t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)
PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
|
| 80 | c2o48X_ |
|
not modelled |
70.6 |
16 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 81 | c3kg2A_ |
|
not modelled |
70.6 |
14 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:glutamate receptor 2;
PDBTitle: ampa subtype ionotropic glutamate receptor in complex with competitive2 antagonist zk 200775
|
| 82 | d1vyra_ |
|
not modelled |
70.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 83 | d1byka_ |
|
not modelled |
69.9 |
12 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 84 | c3m2tA_ |
|
not modelled |
69.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
|
| 85 | c1zq1B_ |
|
not modelled |
68.8 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
|
| 86 | d1xeaa1 |
|
not modelled |
68.3 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 87 | c2fqxA_ |
|
not modelled |
67.9 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:membrane lipoprotein tmpc;
PDBTitle: pnra from treponema pallidum complexed with guanosine
|
| 88 | c3fefB_ |
|
not modelled |
67.8 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative glucosidase lpld;
PDBTitle: crystal structure of putative glucosidase lpld from2 bacillus subtilis
|
| 89 | c2y0fD_ |
|
not modelled |
67.3 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 90 | c3d6nB_ |
|
not modelled |
67.0 |
10 |
PDB header:hydrolase/transferase
Chain: B: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: crystal structure of aquifex dihydroorotase activated by aspartate2 transcarbamoylase
|
| 91 | c3e61A_ |
|
not modelled |
66.7 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative transcriptional repressor of ribose operon;
PDBTitle: crystal structure of a putative transcriptional repressor of ribose2 operon from staphylococcus saprophyticus subsp. saprophyticus
|
| 92 | d1mx3a1 |
|
not modelled |
66.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 93 | d1jaya_ |
|
not modelled |
66.1 |
33 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 94 | c1drwA_ |
|
not modelled |
65.3 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
| 95 | c2q4eB_ |
|
not modelled |
64.6 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
| 96 | c3gr7A_ |
|
not modelled |
64.6 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 97 | c3c52B_ |
|
not modelled |
64.5 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from2 helicobacter pylori in complex with3 phosphoglycolohydroxamic acid, a competitive inhibitor
|
| 98 | c3egcF_ |
|
not modelled |
62.9 |
13 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative ribose operon repressor;
PDBTitle: crystal structure of a putative ribose operon repressor from2 burkholderia thailandensis
|
| 99 | c3q94B_ |
|
not modelled |
62.9 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase, class ii;
PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
|
| 100 | c3jy6B_ |
|
not modelled |
61.9 |
15 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci transcriptional regulator from lactobacillus2 brevis
|
| 101 | d1jkxa_ |
|
not modelled |
61.6 |
20 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 102 | d2d6fa2 |
|
not modelled |
61.6 |
16 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 103 | c3hskB_ |
|
not modelled |
61.6 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 with nadp from candida albicans
|
| 104 | c3qk7C_ |
|
not modelled |
61.3 |
13 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcriptional regulator from yersinia2 pestis biovar microtus str. 91001
|
| 105 | d1i36a2 |
|
not modelled |
60.5 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 106 | c3c3kA_ |
|
not modelled |
60.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of an uncharacterized protein from actinobacillus2 succinogenes
|
| 107 | d2liva_ |
|
not modelled |
59.3 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 108 | d4pfka_ |
|
not modelled |
59.1 |
18 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
| 109 | d1cb0a_ |
|
not modelled |
58.3 |
50 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 110 | d1vc4a_ |
|
not modelled |
58.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 111 | c2ywrA_ |
|
not modelled |
57.8 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of gar transformylase from aquifex2 aeolicus
|
| 112 | d1n1ea2 |
|
not modelled |
57.3 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 113 | c1kjjA_ |
|
not modelled |
56.8 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase 2;
PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
|
| 114 | d1deka_ |
|
not modelled |
56.6 |
45 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 115 | c3e3mA_ |
|
not modelled |
56.5 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of a laci family transcriptional2 regulator from silicibacter pomeroyi
|
| 116 | c3kkeA_ |
|
not modelled |
56.5 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:laci family transcriptional regulator;
PDBTitle: crystal structure of a laci family transcriptional regulator2 from mycobacterium smegmatis
|
| 117 | c3fd8A_ |
|
not modelled |
55.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 118 | c3etjB_ |
|
not modelled |
54.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase atpase
PDBTitle: crystal structure e. coli purk in complex with mg, adp, and2 pi
|
| 119 | c3fa4D_ |
|
not modelled |
54.1 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 120 | d1qcza_ |
|
not modelled |
53.4 |
17 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |