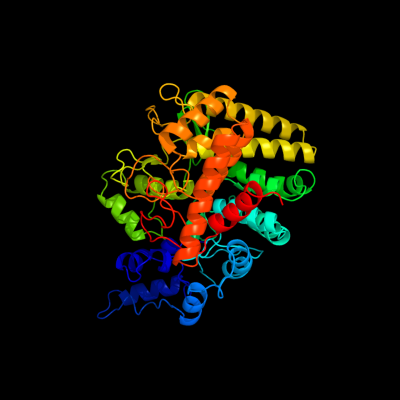
| 1 | d2jg0a1
|
|
 |
100.0 |
51 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Trehalase-like |
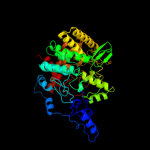
| 2 | c2jg0A_
|
|
 |
100.0 |
51 |
PDB header:hydrolase
Chain: A: PDB Molecule:periplasmic trehalase;
PDBTitle: family 37 trehalase from escherichia coli in complex with 1-2 thiatrehazolin
|
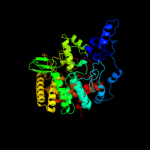
| 3 | c3c67B_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized protein ygjk;
PDBTitle: escherichia coli k12 ygjk in a complexed with glucose
|
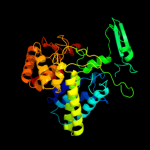
| 4 | c2z07A_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein ttha0978;
PDBTitle: crystal structure of uncharacterized conserved protein from2 thermus thermophilus hb8
|
| 5 | c1v7wA_
|
|
 |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:chitobiose phosphorylase;
PDBTitle: crystal structure of vibrio proteolyticus chitobiose phosphorylase in2 complex with glcnac
|
| 6 | c2okxB_
|
|
 |
99.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:rhamnosidase b;
PDBTitle: crystal structure of gh78 family rhamnosidase of bacillus sp. gl1 at2 1.9 a
|
| 7 | d1v7wa1
|
|
 |
99.9 |
14 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glycosyltransferase family 36 C-terminal domain |
| 8 | c3cihA_
|
|
 |
99.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alpha-rhamnosidase;
PDBTitle: crystal structure of a putative alpha-rhamnosidase from2 bacteroides thetaiotaomicron
|
| 9 | d1lf6a1
|
|
 |
99.8 |
14 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Bacterial glucoamylase C-terminal domain-like |
| 10 | c2cqtA_
|
|
 |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:cellobiose phosphorylase;
PDBTitle: crystal structure of cellvibrio gilvus cellobiose phosphorylase2 crystallized from sodium/potassium phosphate
|
| 11 | c1lf6A_
|
|
 |
99.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucoamylase;
PDBTitle: crystal structure of bacterial glucoamylase
|
| 12 | d1ulva1
|
|
 |
99.3 |
19 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Bacterial glucoamylase C-terminal domain-like |
| 13 | d2fbaa1
|
|
 |
98.8 |
17 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glucoamylase |
| 14 | c2vn4A_
|
|
 |
98.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucoamylase;
PDBTitle: glycoside hydrolase family 15 glucoamylase from hypocrea2 jecorina
|
| 15 | c1ug9A_
|
|
 |
98.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucodextranase;
PDBTitle: crystal structure of glucodextranase from arthrobacter globiformis i42
|
| 16 | d1gaia_
|
|
 |
98.6 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glucoamylase |
| 17 | d2nvpa1
|
|
 |
97.7 |
18 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:CPF0428-like |
| 18 | c2ww1B_
|
|
 |
97.6 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-1,2-mannosidase;
PDBTitle: structure of the family gh92 inverting mannosidase bt39902 from bacteroides thetaiotaomicron vpi-5482 in complex with3 thiomannobioside
|
| 19 | c3qspB_
|
|
 |
97.6 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: analysis of a new family of widely distributed metal-independent alpha2 mannosidases provides unique insight into the processing of n-linked3 glycans, streptococcus pneumoniae sp_2144 non-productive substrate4 complex with alpha-1,6-mannobiose
|
| 20 | c2p0vA_
|
|
 |
97.5 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein bt3781;
PDBTitle: crystal structure of bt3781 protein from bacteroides2 thetaiotaomicron, northeast structural genomics target3 btr58
|
| 21 | d2p0va1 |
|
not modelled |
97.5 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:CPF0428-like |
| 22 | c2eacB_ |
|
not modelled |
97.3 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-fucosidase;
PDBTitle: crystal structure of 1,2-a-l-fucosidase from2 bifidobacterium bifidum in complex with3 deoxyfuconojirimycin
|
| 23 | c2wvyA_ |
|
not modelled |
97.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-1,2-mannosidase;
PDBTitle: structure of the family gh92 inverting mannosidase bt21992 from bacteroides thetaiotaomicron vpi-5482
|
| 24 | d1h54a1 |
|
not modelled |
96.9 |
13 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glycosyltransferase family 36 C-terminal domain |
| 25 | c2rdyB_ |
|
not modelled |
94.6 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:bh0842 protein;
PDBTitle: crystal structure of a putative glycoside hydrolase family2 protein from bacillus halodurans
|
| 26 | c1h54B_ |
|
not modelled |
91.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:maltose phosphorylase;
PDBTitle: maltose phosphorylase from lactobacillus brevis
|
| 27 | c3gt5A_ |
|
not modelled |
85.8 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylglucosamine 2-epimerase;
PDBTitle: crystal structure of an n-acetylglucosamine 2-epimerase family protein2 from xylella fastidiosa
|
| 28 | c3bhwA_ |
|
not modelled |
58.5 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharacterized protein from magnetospirillum2 magneticum
|
| 29 | d1nzpa_ |
|
not modelled |
42.0 |
11 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 30 | d2afaa1 |
|
not modelled |
38.8 |
15 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:N-acylglucosamine (NAG) epimerase |
| 31 | c3k7xA_ |
|
not modelled |
37.1 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin0763 protein;
PDBTitle: crystal structure of the lin0763 protein from listeria2 innocua. northeast structural genomics consortium target3 lkr23.
|
| 32 | c2q07A_ |
|
not modelled |
35.6 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein af0587;
PDBTitle: crystal structure of af0587, a protein of unknown function
|
| 33 | d1q79a3 |
|
not modelled |
33.6 |
12 |
Fold:Ferredoxin-like
Superfamily:PAP/Archaeal CCA-adding enzyme, C-terminal domain
Family:Poly(A) polymerase, PAP, C-terminal domain |
| 34 | c2gz6B_ |
|
not modelled |
29.1 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetyl-d-glucosamine 2-epimerase;
PDBTitle: crystal structure of anabaena sp. ch1 n-acetyl-d-glucosamine 2-2 epimerase at 2.0 a
|
| 35 | c2g2dA_ |
|
not modelled |
28.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:atp:cobalamin adenosyltransferase;
PDBTitle: crystal structure of a putative pduo-type atp:cobalamin2 adenosyltransferase from mycobacterium tuberculosis
|
| 36 | c2qjxA_ |
|
not modelled |
27.9 |
9 |
PDB header:protein binding
Chain: A: PDB Molecule:protein bim1;
PDBTitle: structural basis of microtubule plus end tracking by2 xmap215, clip-170 and eb1
|
| 37 | d1aina_ |
|
not modelled |
26.5 |
12 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 38 | d1wu7a2 |
|
not modelled |
24.5 |
25 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain |
| 39 | d1fp3a_ |
|
not modelled |
21.9 |
15 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:N-acylglucosamine (NAG) epimerase |
| 40 | c1wy1B_ |
|
not modelled |
21.7 |
40 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein ph0671;
PDBTitle: crystal structure of the ph0671 protein from pyrococcus horikoshii ot3
|
| 41 | d2ovra1 |
|
not modelled |
20.8 |
13 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 42 | c1wyoA_ |
|
not modelled |
19.8 |
12 |
PDB header:structural protein
Chain: A: PDB Molecule:microtubule-associated protein rp/eb family
PDBTitle: solution structure of the ch domain of human microtubule-2 associated protein rp/eb family member 3
|
| 43 | d2qjza1 |
|
not modelled |
18.4 |
12 |
Fold:CH domain-like
Superfamily:Calponin-homology domain, CH-domain
Family:Calponin-homology domain, CH-domain |
| 44 | d2bcqa1 |
|
not modelled |
18.3 |
14 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 45 | d1jmsa1 |
|
not modelled |
16.5 |
16 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 46 | c3racA_ |
|
not modelled |
16.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:histidine-trna ligase;
PDBTitle: crystal strucutre of histidine--trna ligase subunit from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446.
|
| 47 | d1dm5a_ |
|
not modelled |
15.1 |
11 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 48 | d2fmpa1 |
|
not modelled |
14.6 |
19 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 49 | c3lhrA_ |
|
not modelled |
14.3 |
19 |
PDB header:transcription regulator
Chain: A: PDB Molecule:zinc finger protein 24;
PDBTitle: crystal structure of the scan domain from human znf24
|
| 50 | d1mcxa_ |
|
not modelled |
13.6 |
12 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 51 | c1wvtA_ |
|
not modelled |
13.6 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein st2180;
PDBTitle: crystal structure of uncharacterized protein st2180 from sulfolobus2 tokodaii
|
| 52 | c3ajfA_ |
|
not modelled |
13.5 |
22 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: structural insigths into dsrna binding and rna silencing suppression2 by ns3 protein of rice hoja blanca tenuivirus
|
| 53 | d1rutx4 |
|
not modelled |
12.7 |
71 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 54 | c2idxA_ |
|
not modelled |
12.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:cob(i)yrinic acid a,c-diamide
PDBTitle: structure of human atp:cobalamin adenosyltransferase bound2 to atp.
|
| 55 | d1iwpa_ |
|
not modelled |
11.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Diol dehydratase, alpha subunit |
| 56 | d1zeta2 |
|
not modelled |
11.2 |
20 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 57 | d1xg7a_ |
|
not modelled |
10.0 |
13 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:AgoG-like |
| 58 | c1kdhA_ |
|
not modelled |
9.8 |
14 |
PDB header:transferase/dna
Chain: A: PDB Molecule:terminal deoxynucleotidyltransferase short
PDBTitle: binary complex of murine terminal deoxynucleotidyl2 transferase with a primer single stranded dna
|
| 59 | c3qsjA_ |
|
not modelled |
9.7 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:nudix hydrolase;
PDBTitle: crystal structure of nudix hydrolase from alicyclobacillus2 acidocaldarius
|
| 60 | d2ie7a1 |
|
not modelled |
9.7 |
13 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 61 | c3lzzB_ |
|
not modelled |
9.7 |
78 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structures of cupin superfamily bbduf985 from branchiostoma2 belcheri tsingtauense in apo and gdp-bound forms
|
| 62 | d1z3xa1 |
|
not modelled |
9.5 |
18 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:GUN4-associated domain |
| 63 | d1jq6a_ |
|
not modelled |
9.1 |
17 |
Fold:Herpes virus serine proteinase, assemblin
Superfamily:Herpes virus serine proteinase, assemblin
Family:Herpes virus serine proteinase, assemblin |
| 64 | d1k8kd1 |
|
not modelled |
8.9 |
28 |
Fold:Secretion chaperone-like
Superfamily:Arp2/3 complex subunits
Family:Arp2/3 complex subunits |
| 65 | d1hvda_ |
|
not modelled |
8.9 |
13 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 66 | c1nj2A_ |
|
not modelled |
8.8 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:proline-trna synthetase;
PDBTitle: crystal structure of prolyl-trna synthetase from methanothermobacter2 thermautotrophicus
|
| 67 | d1avca1 |
|
not modelled |
8.8 |
5 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 68 | c2fllA_ |
|
not modelled |
8.6 |
20 |
PDB header:replication/dna
Chain: A: PDB Molecule:dna polymerase iota;
PDBTitle: ternary complex of human dna polymerase iota with dna and dttp
|
| 69 | c2wseE_ |
|
not modelled |
8.5 |
33 |
PDB header:photosynthesis
Chain: E: PDB Molecule:photosystem i reaction center subunit iv a,
PDBTitle: improved model of plant photosystem i
|
| 70 | c1w3wA_ |
|
not modelled |
8.4 |
10 |
PDB header:coagulation
Chain: A: PDB Molecule:annexin a8;
PDBTitle: the 2.1 angstroem resolution structure of annexin a8
|
| 71 | c2bcuA_ |
|
not modelled |
8.3 |
14 |
PDB header:transferase, lyase/dna
Chain: A: PDB Molecule:dna polymerase lambda;
PDBTitle: dna polymerase lambda in complex with a dna duplex2 containing an unpaired damp and a t:t mismatch
|
| 72 | d1fs1b1 |
|
not modelled |
8.2 |
9 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 73 | d1w7ba_ |
|
not modelled |
8.2 |
5 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 74 | d1m9ia2 |
|
not modelled |
8.1 |
5 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 75 | c3k1qB_ |
|
not modelled |
8.1 |
31 |
PDB header:virus
Chain: B: PDB Molecule:vp3a, the building block protein of inner shell;
PDBTitle: backbone model of an aquareovirus virion by cryo-electron2 microscopy and bioinformatics
|
| 76 | d1alaa_ |
|
not modelled |
8.1 |
13 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 77 | c2doqD_ |
|
not modelled |
7.9 |
27 |
PDB header:cell cycle
Chain: D: PDB Molecule:sfi1p;
PDBTitle: crystal structure of sfi1p/cdc31p complex
|
| 78 | d1nc7a_ |
|
not modelled |
7.8 |
47 |
Fold:Hypothetical protein TM1070
Superfamily:Hypothetical protein TM1070
Family:Hypothetical protein TM1070 |
| 79 | d1dk2a_ |
|
not modelled |
7.7 |
19 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 80 | d1riqa1 |
|
not modelled |
7.5 |
28 |
Fold:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Superfamily:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Family:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS) |
| 81 | d2g4ca2 |
|
not modelled |
7.4 |
22 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain |
| 82 | d1fs2b1 |
|
not modelled |
7.4 |
9 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 83 | c1t94B_ |
|
not modelled |
7.4 |
25 |
PDB header:replication
Chain: B: PDB Molecule:polymerase (dna directed) kappa;
PDBTitle: crystal structure of the catalytic core of human dna2 polymerase kappa
|
| 84 | d1psea_ |
|
not modelled |
7.4 |
55 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Photosystem I accessory protein E (PsaE) |
| 85 | c2lmdA_ |
|
not modelled |
7.3 |
67 |
PDB header:transcription
Chain: A: PDB Molecule:prospero homeobox protein 1;
PDBTitle: minimal constraints solution nmr structure of prospero homeobox2 protein 1 from homo sapiens, northeast structural genomics consortium3 target hr4660b
|
| 86 | c2ah6B_ |
|
not modelled |
7.3 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:bh1595, unknown conserved protein;
PDBTitle: crystal structure of a putative cobalamin adenosyltransferase (bh1595)2 from bacillus halodurans c-125 at 1.60 a resolution
|
| 87 | d1h6za3 |
|
not modelled |
7.2 |
6 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Pyruvate phosphate dikinase, N-terminal domain |
| 88 | d2bgxa1 |
|
not modelled |
7.2 |
29 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:Peptidoglycan binding domain, PGBD |
| 89 | d1z7ma1 |
|
not modelled |
7.2 |
21 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain |
| 90 | c2r7aC_ |
|
not modelled |
7.1 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:bacterial heme binding protein;
PDBTitle: crystal structure of a periplasmic heme binding protein2 from shigella dysenteriae
|
| 91 | c3pg8B_ |
|
not modelled |
7.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
| 92 | d1gxie_ |
|
not modelled |
7.0 |
48 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Photosystem I accessory protein E (PsaE) |
| 93 | c1t3nB_ |
|
not modelled |
7.0 |
21 |
PDB header:replication/dna
Chain: B: PDB Molecule:polymerase (dna directed) iota;
PDBTitle: structure of the catalytic core of dna polymerase iota in2 complex with dna and dttp
|
| 94 | c3k1qC_ |
|
not modelled |
7.0 |
31 |
PDB header:virus
Chain: C: PDB Molecule:vp3b, the building block protein of inner shell;
PDBTitle: backbone model of an aquareovirus virion by cryo-electron2 microscopy and bioinformatics
|
| 95 | d2q07a2 |
|
not modelled |
6.9 |
17 |
Fold:Uracil-DNA glycosylase-like
Superfamily:Uracil-DNA glycosylase-like
Family:AF0587 domain-like |
| 96 | d1m9ia1 |
|
not modelled |
6.9 |
5 |
Fold:Annexin
Superfamily:Annexin
Family:Annexin |
| 97 | d1usya_ |
|
not modelled |
6.9 |
21 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain |
| 98 | d1r8ea2 |
|
not modelled |
6.7 |
14 |
Fold:Probable bacterial effector-binding domain
Superfamily:Probable bacterial effector-binding domain
Family:Multidrug-binding domain of transcription activator BmrR |
| 99 | d1cfra_ |
|
not modelled |
6.5 |
17 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Cfr10I/Bse634I |