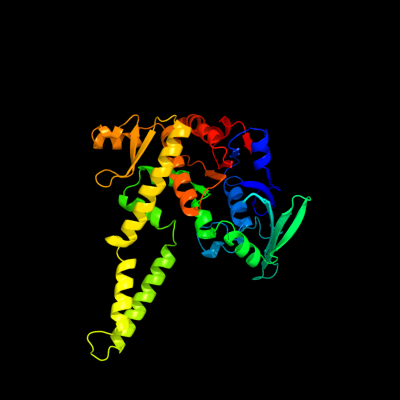
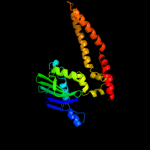
1 c3qg5A_
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: rad50;PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
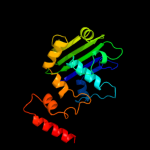
2 c1ii8A_
99.9
20
PDB header: replicationChain: A: PDB Molecule: rad50 abc-atpase;PDBTitle: crystal structure of the p. furiosus rad50 atpase domain
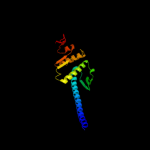
3 c1xexA_
99.9
19
PDB header: cell cycleChain: A: PDB Molecule: smc protein;PDBTitle: structural biochemistry of atp-driven dimerization and dna2 stimulated activation of smc atpases.
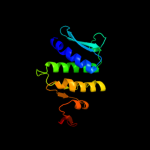
4 c3auyB_
99.9
22
PDB header: recombinationChain: B: PDB Molecule: dna double-strand break repair rad50 atpase;PDBTitle: crystal structure of rad50 bound to adp
5 c3qkuB_
99.9
21
PDB header: replicationChain: B: PDB Molecule: dna double-strand break repair rad50 atpase;PDBTitle: mre11 rad50 binding domain in complex with rad50 and amp-pnp
6 c1ii8B_
99.9
22
PDB header: replicationChain: B: PDB Molecule: rad50 abc-atpase;PDBTitle: crystal structure of the p. furiosus rad50 atpase domain
7 c1f2uD_
99.8
26
PDB header: replicationChain: D: PDB Molecule: rad50 abc-atpase;PDBTitle: crystal structure of rad50 abc-atpase
8 d1e69a_
99.8
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like9 c1us8A_
99.8
29
PDB header: dna repairChain: A: PDB Molecule: dna double-strand break repair rad50 atpase;PDBTitle: the rad50 signature motif: essential to atp binding and2 biological function
10 c1xexB_
99.7
19
PDB header: cell cycleChain: B: PDB Molecule: smc protein;PDBTitle: structural biochemistry of atp-driven dimerization and dna2 stimulated activation of smc atpases.
11 d1w1wa_
99.7
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like12 c2o5vA_
99.6
26
PDB header: replication/recombinationChain: A: PDB Molecule: dna replication and repair protein recf;PDBTitle: recombination mediator recf
13 d1qhla_
99.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like14 c3pihA_
99.2
26
PDB header: hydrolase/dnaChain: A: PDB Molecule: uvrabc system protein a;PDBTitle: t. maritima uvra in complex with fluorescein-modified dna
15 c3bk7A_
99.2
25
PDB header: hydrolyase/translationChain: A: PDB Molecule: abc transporter atp-binding protein;PDBTitle: structure of the complete abce1/rnaase-l inhibitor protein2 from pyrococcus abysii
16 c1yqtA_
99.2
30
PDB header: hydrolyase/translationChain: A: PDB Molecule: rnase l inhibitor;PDBTitle: rnase-l inhibitor
17 c2vf7B_
99.2
24
PDB header: dna-binding proteinChain: B: PDB Molecule: excinuclease abc, subunit a.;PDBTitle: crystal structure of uvra2 from deinococcus radiodurans
18 c2ygrD_
99.1
30
PDB header: hydrolaseChain: D: PDB Molecule: uvrabc system protein a;PDBTitle: mycobacterium tuberculosis uvra
19 c3ozxA_
99.0
20
PDB header: hydrolase, translationChain: A: PDB Molecule: rnase l inhibitor;PDBTitle: crystal structure of abce1 of sulfolubus solfataricus (-fes domain)
20 c2pcjB_
98.9
33
PDB header: hydrolaseChain: B: PDB Molecule: lipoprotein-releasing system atp-binding protein lold;PDBTitle: crystal structure of abc transporter (aq_297) from aquifex aeolicus2 vf5
21 c3dhwC_
not modelled
98.9
35
PDB header: membrane protein/hydrolaseChain: C: PDB Molecule: methionine import atp-binding protein metn;PDBTitle: crystal structure of methionine importer metni
22 c2yl4A_
not modelled
98.8
30
PDB header: membrane proteinChain: A: PDB Molecule: atp-binding cassette sub-family b member 10,PDBTitle: structure of the human mitochondrial abc transporter, abcb10
23 d1g6ha_
not modelled
98.8
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like24 d1b0ua_
not modelled
98.8
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like25 c2olkD_
not modelled
98.8
25
PDB header: hydrolaseChain: D: PDB Molecule: amino acid abc transporter;PDBTitle: abc protein artp in complex with adp-beta-s
26 c2nq2C_
not modelled
98.8
29
PDB header: metal transportChain: C: PDB Molecule: hypothetical abc transporter atp-binding proteinPDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
27 c1vciA_
not modelled
98.8
23
PDB header: transport proteinChain: A: PDB Molecule: sugar-binding transport atp-binding protein;PDBTitle: crystal structure of the atp-binding cassette of multisugar2 transporter from pyrococcus horikoshii ot3 complexed with3 atp
28 c2iw3B_
not modelled
98.8
24
PDB header: translationChain: B: PDB Molecule: elongation factor 3a;PDBTitle: elongation factor 3 in complex with adp
29 c2d62A_
not modelled
98.8
26
PDB header: sugar binding proteinChain: A: PDB Molecule: multiple sugar-binding transport atp-bindingPDBTitle: crystal structure of multiple sugar binding transport atp-2 binding protein
30 d1vpla_
not modelled
98.8
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like31 c3fvqB_
not modelled
98.8
32
PDB header: hydrolaseChain: B: PDB Molecule: fe(3+) ions import atp-binding protein fbpc;PDBTitle: crystal structure of the nucleotide binding domain fbpc2 complexed with atp
32 c2yz2B_
not modelled
98.8
31
PDB header: hydrolaseChain: B: PDB Molecule: putative abc transporter atp-binding protein tm_0222;PDBTitle: crystal structure of the abc transporter in the cobalt transport2 system
33 c1oxtB_
not modelled
98.8
35
PDB header: transport proteinChain: B: PDB Molecule: abc transporter, atp binding protein;PDBTitle: crystal structure of glcv, the abc-atpase of the glucose abc2 transporter from sulfolobus solfataricus
34 c2hydB_
not modelled
98.8
28
PDB header: transport proteinChain: B: PDB Molecule: abc transporter homolog;PDBTitle: multidrug abc transporter sav1866
35 d1l2ta_
not modelled
98.7
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like36 d1v43a3
not modelled
98.7
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like37 d2hyda1
not modelled
98.7
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like38 c3eukC_
not modelled
98.7
19
PDB header: cell cycleChain: C: PDB Molecule: chromosome partition protein mukb, linker;PDBTitle: crystal structure of muke-mukf(residues 292-443)-mukb(head2 domain)-atpgammas complex, asymmetric dimer
39 d3dhwc1
not modelled
98.7
35
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like40 c2pjzA_
not modelled
98.7
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein st1066;PDBTitle: the crystal structure of putative cobalt transport atp-2 binding protein (cbio-2), st1066
41 c2yyzA_
not modelled
98.7
28
PDB header: transport proteinChain: A: PDB Molecule: sugar abc transporter, atp-binding protein;PDBTitle: crystal structure of sugar abc transporter, atp-binding protein
42 c3gfoA_
not modelled
98.7
25
PDB header: atp binding proteinChain: A: PDB Molecule: cobalt import atp-binding protein cbio 1;PDBTitle: structure of cbio1 from clostridium perfringens: part of2 the abc transporter complex cbionq.
43 c2d2fA_
not modelled
98.7
21
PDB header: protein bindingChain: A: PDB Molecule: sufc protein;PDBTitle: crystal structure of atypical cytoplasmic abc-atpase sufc from thermus2 thermophilus hb8
44 d1g2912
not modelled
98.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like45 d3b60a1
not modelled
98.7
31
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like46 d1jj7a_
not modelled
98.7
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like47 d1ji0a_
not modelled
98.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like48 d2pmka1
not modelled
98.7
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like49 c1z47B_
not modelled
98.7
26
PDB header: ligand binding proteinChain: B: PDB Molecule: putative abc-transporter atp-binding protein;PDBTitle: structure of the atpase subunit cysa of the putative2 sulfate atp-binding cassette (abc) transporter from3 alicyclobacillus acidocaldarius
50 c2it1B_
not modelled
98.7
30
PDB header: transport proteinChain: B: PDB Molecule: 362aa long hypothetical maltose/maltodextrinPDBTitle: structure of ph0203 protein from pyrococcus horikoshii
51 c2r6fA_
not modelled
98.7
31
PDB header: hydrolaseChain: A: PDB Molecule: excinuclease abc subunit a;PDBTitle: crystal structure of bacillus stearothermophilus uvra
52 d1mv5a_
not modelled
98.6
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like53 d3d31a2
not modelled
98.6
36
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like54 c2cbzA_
not modelled
98.6
24
PDB header: transportChain: A: PDB Molecule: multidrug resistance-associated protein 1;PDBTitle: structure of the human multidrug resistance protein 12 nucleotide binding domain 1
55 d1oxxk2
not modelled
98.6
33
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like56 d1r0wa_
not modelled
98.6
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like57 c3g5uB_
not modelled
98.6
32
PDB header: membrane proteinChain: B: PDB Molecule: multidrug resistance protein 1a;PDBTitle: structure of p-glycoprotein reveals a molecular basis for2 poly-specific drug binding
58 c3b5xB_
not modelled
98.6
30
PDB header: membrane proteinChain: B: PDB Molecule: lipid a export atp-binding/permease protein msba;PDBTitle: crystal structure of msba from vibrio cholerae
59 c3b5wE_
not modelled
98.6
31
PDB header: membrane proteinChain: E: PDB Molecule: lipid a export atp-binding/permease protein msba;PDBTitle: crystal structure of eschericia coli msba
60 c3d31B_
not modelled
98.6
36
PDB header: transport proteinChain: B: PDB Molecule: sulfate/molybdate abc transporter, atp-bindingPDBTitle: modbc from methanosarcina acetivorans
61 c3nhaA_
not modelled
98.6
25
PDB header: transport proteinChain: A: PDB Molecule: atp-binding cassette sub-family b member 6, mitochondrial;PDBTitle: nucleotide binding domain of human abcb6 (adp mg bound structure)
62 d1xmia_
not modelled
98.6
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like63 c2pzfB_
not modelled
98.5
26
PDB header: hydrolaseChain: B: PDB Molecule: cystic fibrosis transmembrane conductance regulator;PDBTitle: minimal human cftr first nucleotide binding domain as a head-to-tail2 dimer with delta f508
64 d2onka1
not modelled
98.5
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like65 d1l7vc_
not modelled
98.5
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like66 c2d3wB_
not modelled
98.5
20
PDB header: biosynthetic proteinChain: B: PDB Molecule: probable atp-dependent transporter sufc;PDBTitle: crystal structure of escherichia coli sufc, an atpase2 compenent of the suf iron-sulfur cluster assembly machinery
67 c1q1bD_
not modelled
98.5
22
PDB header: transport proteinChain: D: PDB Molecule: maltose/maltodextrin transport atp-binding protein malk;PDBTitle: crystal structure of e. coli malk in the nucleotide-free form
68 d1pf4a1
not modelled
98.4
33
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like69 c2ihyB_
not modelled
98.3
29
PDB header: hydrolaseChain: B: PDB Molecule: abc transporter, atp-binding protein;PDBTitle: structure of the staphylococcus aureus putative atpase subunit of an2 atp-binding cassette (abc) transporter
70 c3gd7C_
not modelled
98.3
24
PDB header: hydrolaseChain: C: PDB Molecule: fusion complex of cystic fibrosis transmembranePDBTitle: crystal structure of human nbd2 complexed with n6-2 phenylethyl-atp (p-atp)
71 d1sgwa_
not modelled
98.2
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like72 c2ghiD_
not modelled
98.0
25
PDB header: transport proteinChain: D: PDB Molecule: transport protein;PDBTitle: crystal structure of plasmodium yoelii multidrug resistance2 protein 2
73 d2awna2
not modelled
97.9
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like74 d1vmaa2
not modelled
97.7
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like75 d1np6a_
not modelled
97.6
43
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like76 d2qy9a2
not modelled
97.6
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like77 d1okkd2
not modelled
97.5
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like78 c2j41A_
not modelled
97.5
40
PDB header: transferaseChain: A: PDB Molecule: guanylate kinase;PDBTitle: crystal structure of staphylococcus aureus guanylate2 monophosphate kinase
79 c3tauB_
not modelled
97.4
40
PDB header: transferaseChain: B: PDB Molecule: guanylate kinase;PDBTitle: crystal structure of a putative guanylate monophosphaste kinase from2 listeria monocytogenes egd-e
80 d1ls1a2
not modelled
97.4
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like81 c3a4mB_
not modelled
97.3
35
PDB header: transferaseChain: B: PDB Molecule: l-seryl-trna(sec) kinase;PDBTitle: crystal structure of archaeal o-phosphoseryl-trna(sec)2 kinase
82 d1qzxa3
not modelled
97.1
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like83 d2i3ba1
not modelled
97.0
47
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)84 d1ye8a1
not modelled
97.0
53
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)85 d1cr2a_
not modelled
96.9
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)86 d1ewqa2
not modelled
96.9
41
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like87 d1znwa1
not modelled
96.7
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases88 c1ewqA_
not modelled
96.7
44
PDB header: replication/dnaChain: A: PDB Molecule: dna mismatch repair protein muts;PDBTitle: crystal structure taq muts complexed with a heteroduplex2 dna at 2.2 a resolution
89 d1wb9a2
not modelled
96.7
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like90 c2o8bA_
not modelled
96.6
27
PDB header: dna binding protein/dnaChain: A: PDB Molecule: dna mismatch repair protein msh2;PDBTitle: human mutsalpha (msh2/msh6) bound to adp and a g t mispair
91 c2o8dB_
not modelled
96.6
31
PDB header: dna binding protein/dnaChain: B: PDB Molecule: dna mismatch repair protein msh6;PDBTitle: human mutsalpha (msh2/msh6) bound to adp and a g du mispair
92 c3jvvA_
not modelled
96.6
50
PDB header: atp binding proteinChain: A: PDB Molecule: twitching mobility protein;PDBTitle: crystal structure of p. aeruginosa pilt with bound amp-pcp
93 d1ki9a_
not modelled
96.6
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases94 d1p9ra_
not modelled
96.6
48
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)95 c2cnwF_
not modelled
96.6
18
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
96 d1s96a_
not modelled
96.6
40
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases97 c2j7pA_
not modelled
96.5
24
PDB header: signal recognitionChain: A: PDB Molecule: signal recognition particle protein;PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
98 c2yhsA_
not modelled
96.5
18
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the e. coli srp receptor ftsy
99 c3thxB_
not modelled
96.5
31
PDB header: dna binding protein/dnaChain: B: PDB Molecule: dna mismatch repair protein msh3;PDBTitle: human mutsbeta complexed with an idl of 3 bases (loop3) and adp
100 c3b9qA_
not modelled
96.5
30
PDB header: protein transportChain: A: PDB Molecule: chloroplast srp receptor homolog, alpha subunitPDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
101 c1u9iA_
not modelled
96.5
19
PDB header: circadian clock proteinChain: A: PDB Molecule: kaic;PDBTitle: crystal structure of circadian clock protein kaic with phosphorylation2 sites
102 c3geiB_
not modelled
96.5
44
PDB header: hydrolaseChain: B: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from chlorobium tepidum in complex2 with gcp
103 d1zp6a1
not modelled
96.4
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Atu3015-like104 c1wbdA_
not modelled
96.4
31
PDB header: dna-bindingChain: A: PDB Molecule: dna mismatch repair protein muts;PDBTitle: crystal structure of e. coli dna mismatch repair enzyme2 muts, e38q mutant, in complex with a g.t mismatch
105 c2og2A_
not modelled
96.4
30
PDB header: protein transportChain: A: PDB Molecule: putative signal recognition particle receptor;PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
106 d1sxje2
not modelled
96.4
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain107 c2f1rA_
not modelled
96.4
35
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin-guanine dinucleotide biosynthesisPDBTitle: crystal structure of molybdopterin-guanine biosynthesis2 protein b (mobb)
108 c1ewrA_
not modelled
96.4
44
PDB header: hydrolaseChain: A: PDB Molecule: dna mismatch repair protein muts;PDBTitle: crystal structure of taq muts
109 c2qy9A_
not modelled
96.4
18
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
110 d1sxja2
not modelled
96.4
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain111 d1yrba1
not modelled
96.3
35
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like112 c1znyA_
not modelled
96.3
25
PDB header: transferaseChain: A: PDB Molecule: guanylate kinase;PDBTitle: crystal structure of mycobacterium tuberculosis guanylate kinase in2 complex with gdp
113 c2eyuA_
not modelled
96.3
38
PDB header: protein transportChain: A: PDB Molecule: twitching motility protein pilt;PDBTitle: the crystal structure of the c-terminal domain of aquifex2 aeolicus pilt
114 c1wxqA_
not modelled
96.3
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of gtp binding protein from pyrococcus horikoshii2 ot3
115 c2gzaB_
not modelled
96.2
39
PDB header: hydrolaseChain: B: PDB Molecule: type iv secretion system protein virb11;PDBTitle: crystal structure of the virb11 atpase from the brucella suis type iv2 secretion system in complex with sulphate
116 c1zu4A_
not modelled
96.2
24
PDB header: protein transportChain: A: PDB Molecule: ftsy;PDBTitle: crystal structure of ftsy from mycoplasma mycoides- space2 group p21212
117 d1g6oa_
not modelled
96.2
38
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)118 c3neyC_
not modelled
96.2
20
PDB header: membrane proteinChain: C: PDB Molecule: 55 kda erythrocyte membrane protein;PDBTitle: crystal structure of the kinase domain of mpp1/p55
119 c2gszE_
not modelled
96.2
43
PDB header: protein transportChain: E: PDB Molecule: twitching motility protein pilt;PDBTitle: structure of a. aeolicus pilt with 6 monomers per2 asymmetric unit
120 c1vmaA_
not modelled
96.1
21
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution