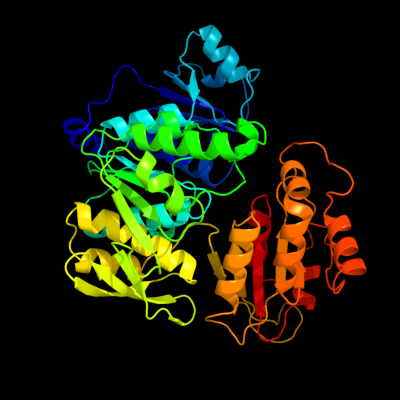
| 1 | c2f00A_
|
|
 |
100.0 |
96 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: escherichia coli murc
|
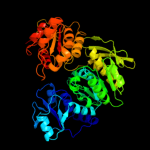
| 2 | c1j6uA_
|
|
 |
100.0 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase2 murc (tm0231) from thermotoga maritima at 2.3 a resolution
|
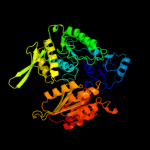
| 3 | c1gqqA_
|
|
 |
100.0 |
65 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus2 influenzae
|
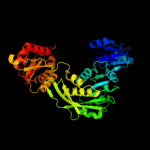
| 4 | c3hn7A_
|
|
 |
100.0 |
26 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: crystal structure of a murein peptide ligase mpl (psyc_0032) from2 psychrobacter arcticus 273-4 at 1.65 a resolution
|
| 5 | c3uagA_
|
|
 |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:protein (udp-n-acetylmuramoyl-l-alanine:d-
PDBTitle: udp-n-acetylmuramoyl-l-alanine:d-glutamate ligase
|
| 6 | c3lk7A_
|
|
 |
100.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: the crystal structure of udp-n-acetylmuramoylalanine-d-2 glutamate (murd) ligase from streptococcus agalactiae to3 1.5a
|
| 7 | c3eagA_
|
|
 |
100.0 |
30 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-
PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
|
| 8 | c1e8cB_
|
|
 |
100.0 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamate--2,6-
PDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide2 synthetase from e. coli
|
| 9 | c2vosA_
|
|
 |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:folylpolyglutamate synthase protein folc;
PDBTitle: mycobacterium tuberculosis folylpolyglutamate synthase2 complexed with adp
|
| 10 | c2wtzC_
|
|
 |
100.0 |
24 |
PDB header:ligase
Chain: C: PDB Molecule:udp-n-acetylmuramoyl-l-alanyl-d-glutamate-
PDBTitle: mure ligase of mycobacterium tuberculosis
|
| 11 | c2am1A_
|
|
 |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine-d-glutamyl-lysine-d-alanyl-d-
PDBTitle: sp protein ligand 1
|
| 12 | c1gg4A_
|
|
 |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamyl-2,6-
PDBTitle: crystal structure of escherichia coli udpmurnac-tripeptide2 d-alanyl-d-alanine-adding enzyme (murf) at 2.3 angstrom3 resolution
|
| 13 | c1w78A_
|
|
 |
100.0 |
20 |
PDB header:synthase
Chain: A: PDB Molecule:folc bifunctional protein;
PDBTitle: e.coli folc in complex with dhpp and adp
|
| 14 | c1o5zA_
|
|
 |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:folylpolyglutamate synthase/dihydrofolate synthase;
PDBTitle: crystal structure of folylpolyglutamate synthase (tm0166) from2 thermotoga maritima at 2.10 a resolution
|
| 15 | c2gc6A_
|
|
 |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:folylpolyglutamate synthase;
PDBTitle: s73a mutant of l. casei fpgs
|
| 16 | c3n2aA_
|
|
 |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:bifunctional folylpolyglutamate synthase/dihydrofolate
PDBTitle: crystal structure of bifunctional folylpolyglutamate2 synthase/dihydrofolate synthase from yersinia pestis co92
|
| 17 | d1p3da3
|
|
 |
100.0 |
69 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:MurCDEF |
| 18 | d1j6ua3
|
|
 |
100.0 |
26 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:MurCDEF |
| 19 | d2jfga3
|
|
 |
100.0 |
22 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:MurCDEF |
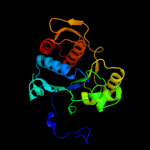
| 20 | d2gc6a2
|
|
 |
100.0 |
17 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:Folylpolyglutamate synthetase |
| 21 | d1e8ca3 |
|
not modelled |
100.0 |
18 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:MurCDEF |
| 22 | d1gg4a4 |
|
not modelled |
100.0 |
22 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:MurCDEF |
| 23 | d1o5za2 |
|
not modelled |
100.0 |
17 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:Folylpolyglutamate synthetase |
| 24 | d1p3da2 |
|
not modelled |
99.9 |
62 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 25 | d1p3da1 |
|
not modelled |
99.9 |
64 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 26 | d1j6ua1 |
|
not modelled |
99.9 |
36 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 27 | d1j6ua2 |
|
not modelled |
99.8 |
26 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 28 | d1e8ca2 |
|
not modelled |
99.8 |
19 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 29 | c3mvnA_ |
|
not modelled |
99.8 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
| 30 | d2jfga1 |
|
not modelled |
99.7 |
18 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 31 | d1o5za1 |
|
not modelled |
99.7 |
15 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:Folylpolyglutamate synthetase, C-terminal domain |
| 32 | d2jfga2 |
|
not modelled |
99.7 |
15 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 33 | d1gg4a1 |
|
not modelled |
99.6 |
16 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 34 | d2gc6a1 |
|
not modelled |
99.3 |
13 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:Folylpolyglutamate synthetase, C-terminal domain |
| 35 | d1pjqa1 |
|
not modelled |
98.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 36 | c3cumA_ |
|
not modelled |
98.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
| 37 | c3g79A_ |
|
not modelled |
97.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ndp-n-acetyl-d-galactosaminuronic acid dehydrogenase;
PDBTitle: crystal structure of ndp-n-acetyl-d-galactosaminuronic acid2 dehydrogenase from methanosarcina mazei go1
|
| 38 | d1uxja1 |
|
not modelled |
97.9 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 39 | c2o3jC_ |
|
not modelled |
97.8 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of caenorhabditis elegans udp-glucose dehydrogenase
|
| 40 | c3gg2B_ |
|
not modelled |
97.7 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sugar dehydrogenase, udp-glucose/gdp-mannose
PDBTitle: crystal structure of udp-glucose 6-dehydrogenase from2 porphyromonas gingivalis bound to product udp-glucuronate
|
| 41 | c3hn2A_ |
|
not modelled |
97.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of 2-dehydropantoate 2-reductase from geobacter2 metallireducens gs-15
|
| 42 | d3cuma2 |
|
not modelled |
97.7 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 43 | c2g5cD_ |
|
not modelled |
97.7 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
|
| 44 | c1vpdA_ |
|
not modelled |
97.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tartronate semialdehyde reductase;
PDBTitle: x-ray crystal structure of tartronate semialdehyde reductase2 [salmonella typhimurium lt2]
|
| 45 | c2y0dB_ |
|
not modelled |
97.6 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: bcec mutation y10k
|
| 46 | c3k96B_ |
|
not modelled |
97.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
| 47 | c3ckyA_ |
|
not modelled |
97.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-hydroxymethyl glutarate dehydrogenase;
PDBTitle: structural and kinetic properties of a beta-hydroxyacid dehydrogenase2 involved in nicotinate fermentation
|
| 48 | c3d4oA_ |
|
not modelled |
97.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 49 | c3ojlA_ |
|
not modelled |
97.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cap5o;
PDBTitle: native structure of the udp-n-acetyl-mannosamine dehydrogenase cap5o2 from staphylococcus aureus
|
| 50 | c2hk8B_ |
|
not modelled |
97.6 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
|
| 51 | c1ps9A_ |
|
not modelled |
97.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 52 | c3ghyA_ |
|
not modelled |
97.6 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ketopantoate reductase protein;
PDBTitle: crystal structure of a putative ketopantoate reductase from ralstonia2 solanacearum molk2
|
| 53 | c3l6dB_ |
|
not modelled |
97.6 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
| 54 | c3pduF_ |
|
not modelled |
97.6 |
22 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:3-hydroxyisobutyrate dehydrogenase family protein;
PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter sulfurreducens in complex with nadp+
|
| 55 | c3d1lB_ |
|
not modelled |
97.6 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
| 56 | c1pjtB_ |
|
not modelled |
97.5 |
20 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg,2 the multifunctional3 methyltransferase/dehydrogenase/ferrochelatase for4 siroheme synthesis
|
| 57 | c1bg6A_ |
|
not modelled |
97.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;
PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
|
| 58 | c2pv7B_ |
|
not modelled |
97.5 |
32 |
PDB header:isomerase, oxidoreductase
Chain: B: PDB Molecule:t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)
PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
|
| 59 | c3ktdC_ |
|
not modelled |
97.5 |
12 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
| 60 | c3qhaB_ |
|
not modelled |
97.5 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
|
| 61 | c3prjB_ |
|
not modelled |
97.5 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: role of packing defects in the evolution of allostery and induced fit2 in human udp-glucose dehydrogenase.
|
| 62 | d1l7da1 |
|
not modelled |
97.5 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 63 | d1n1ea2 |
|
not modelled |
97.5 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 64 | c2ev9B_ |
|
not modelled |
97.4 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from thermus2 thermophilus hb8 in complex with nadp(h) and shikimate
|
| 65 | c1mv8A_ |
|
not modelled |
97.4 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gdp-mannose 6-dehydrogenase;
PDBTitle: 1.55 a crystal structure of a ternary complex of gdp-mannose2 dehydrogenase from psuedomonas aeruginosa
|
| 66 | d1e5qa1 |
|
not modelled |
97.4 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 67 | c3dfzB_ |
|
not modelled |
97.4 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
| 68 | d1bg6a2 |
|
not modelled |
97.4 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 69 | c2q3eH_ |
|
not modelled |
97.4 |
15 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of human udp-glucose dehydrogenase complexed with nadh and2 udp-glucose
|
| 70 | d1mv8a2 |
|
not modelled |
97.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 71 | d2f1ka2 |
|
not modelled |
97.4 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 72 | c2f1kD_ |
|
not modelled |
97.4 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
|
| 73 | d1vpda2 |
|
not modelled |
97.4 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 74 | c1np3B_ |
|
not modelled |
97.4 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 75 | c3hwrA_ |
|
not modelled |
97.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of pane/apba family ketopantoate reductase2 (yp_299159.1) from ralstonia eutropha jmp134 at 2.15 a resolution
|
| 76 | d1a4ia1 |
|
not modelled |
97.4 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 77 | d1nyta1 |
|
not modelled |
97.4 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 78 | c3pefA_ |
|
not modelled |
97.4 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, nad-binding;
PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter metallireducens in complex with nadp+
|
| 79 | d1ez4a1 |
|
not modelled |
97.4 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 80 | c3ggpA_ |
|
not modelled |
97.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from a. aeolicus in2 complex with hydroxyphenyl propionate and nad+
|
| 81 | c1djnB_ |
|
not modelled |
97.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trimethylamine dehydrogenase;
PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
|
| 82 | c3k30B_ |
|
not modelled |
97.3 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
| 83 | c1gthD_ |
|
not modelled |
97.3 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydropyrimidine dehydrogenase;
PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
|
| 84 | c1npyA_ |
|
not modelled |
97.3 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical shikimate 5-dehydrogenase-like
PDBTitle: structure of shikimate 5-dehydrogenase-like protein hi0607
|
| 85 | d9ldta1 |
|
not modelled |
97.3 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 86 | c1z82A_ |
|
not modelled |
97.3 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glycerol-3-phosphate dehydrogenase (tm0378) from2 thermotoga maritima at 2.00 a resolution
|
| 87 | c2uyyD_ |
|
not modelled |
97.3 |
18 |
PDB header:cytokine
Chain: D: PDB Molecule:n-pac protein;
PDBTitle: structure of the cytokine-like nuclear factor n-pac
|
| 88 | c3c24A_ |
|
not modelled |
97.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase (yp_511008.1) from2 jannaschia sp. ccs1 at 1.62 a resolution
|
| 89 | c2rirA_ |
|
not modelled |
97.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 90 | c3dojA_ |
|
not modelled |
97.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase-like protein;
PDBTitle: structure of glyoxylate reductase 1 from arabidopsis2 (atglyr1)
|
| 91 | c3g0oA_ |
|
not modelled |
97.2 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of 3-hydroxyisobutyrate dehydrogenase2 (ygbj) from salmonella typhimurium
|
| 92 | d2cvza2 |
|
not modelled |
97.2 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 93 | c1i36A_ |
|
not modelled |
97.2 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein mth1747;
PDBTitle: structure of conserved protein mth1747 of unknown function2 reveals structural similarity with 3-hydroxyacid3 dehydrogenases
|
| 94 | d1f0ya2 |
|
not modelled |
97.2 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 95 | d1guza1 |
|
not modelled |
97.2 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 96 | c3plnA_ |
|
not modelled |
97.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: crystal structure of klebsiella pneumoniae udp-glucose 6-dehydrogenase2 complexed with udp-glucose
|
| 97 | d1pzga1 |
|
not modelled |
97.2 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 98 | d1np3a2 |
|
not modelled |
97.1 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 99 | c2eggA_ |
|
not modelled |
97.1 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from2 geobacillus kaustophilus
|
| 100 | c1e5lA_ |
|
not modelled |
97.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
| 101 | d2ahra2 |
|
not modelled |
97.1 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 102 | c3triB_ |
|
not modelled |
97.1 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: structure of a pyrroline-5-carboxylate reductase (proc) from coxiella2 burnetii
|
| 103 | c1nytC_ |
|
not modelled |
97.1 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: shikimate dehydrogenase aroe complexed with nadp+
|
| 104 | d1i36a2 |
|
not modelled |
97.1 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 105 | d1p77a1 |
|
not modelled |
97.1 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 106 | c2ew2B_ |
|
not modelled |
97.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase, putative;
PDBTitle: crystal structure of the putative 2-dehydropantoate 2-reductase from2 enterococcus faecalis
|
| 107 | c3o8qB_ |
|
not modelled |
97.1 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase i alpha;
PDBTitle: 1.45 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae
|
| 108 | d1b0aa1 |
|
not modelled |
97.1 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 109 | c2ag8A_ |
|
not modelled |
97.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: nadp complex of pyrroline-5-carboxylate reductase from neisseria2 meningitidis
|
| 110 | d1ldma1 |
|
not modelled |
97.1 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 111 | c1m67A_ |
|
not modelled |
97.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of leishmania mexicana gpdh complexed with inhibitor2 2-bromo-6-hydroxy-purine
|
| 112 | c2ahrB_ |
|
not modelled |
97.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative pyrroline carboxylate reductase;
PDBTitle: crystal structures of 1-pyrroline-5-carboxylate reductase from human2 pathogen streptococcus pyogenes
|
| 113 | d1li4a1 |
|
not modelled |
97.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 114 | c2cvzD_ |
|
not modelled |
97.0 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: structure of hydroxyisobutyrate dehydrogenase from thermus2 thermophilus hb8
|
| 115 | d1i10a1 |
|
not modelled |
97.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 116 | c2o7qA_ |
|
not modelled |
97.0 |
17 |
PDB header:oxidoreductase,transferase
Chain: A: PDB Molecule:bifunctional 3-dehydroquinate dehydratase/shikimate
PDBTitle: crystal structure of the a. thaliana dhq-dehydroshikimate-sdh-2 shikimate-nadp(h)
|
| 117 | c3eywA_ |
|
not modelled |
97.0 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 118 | c3gviB_ |
|
not modelled |
97.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of lactate/malate dehydrogenase from2 brucella melitensis in complex with adp
|
| 119 | d1c1da1 |
|
not modelled |
97.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 120 | c3ic5A_ |
|
not modelled |
97.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|