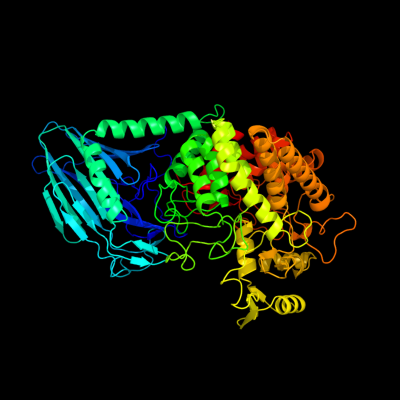
| 1 | c3c67B_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized protein ygjk;
PDBTitle: escherichia coli k12 ygjk in a complexed with glucose
|
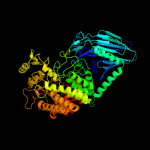
| 2 | d2jg0a1
|
|
 |
100.0 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Trehalase-like |
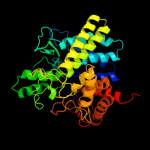
| 3 | c2jg0A_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:periplasmic trehalase;
PDBTitle: family 37 trehalase from escherichia coli in complex with 1-2 thiatrehazolin
|
| 4 | c2z07A_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein ttha0978;
PDBTitle: crystal structure of uncharacterized conserved protein from2 thermus thermophilus hb8
|
| 5 | c1v7wA_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:chitobiose phosphorylase;
PDBTitle: crystal structure of vibrio proteolyticus chitobiose phosphorylase in2 complex with glcnac
|
| 6 | c2cqtA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:cellobiose phosphorylase;
PDBTitle: crystal structure of cellvibrio gilvus cellobiose phosphorylase2 crystallized from sodium/potassium phosphate
|
| 7 | d1v7wa1
|
|
 |
100.0 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glycosyltransferase family 36 C-terminal domain |
| 8 | d1lf6a1
|
|
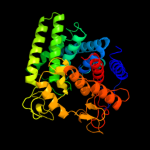 |
100.0 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Bacterial glucoamylase C-terminal domain-like |
| 9 | c2okxB_
|
|
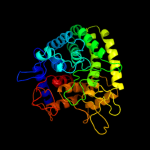 |
99.9 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:rhamnosidase b;
PDBTitle: crystal structure of gh78 family rhamnosidase of bacillus sp. gl1 at2 1.9 a
|
| 10 | c1lf6A_
|
|
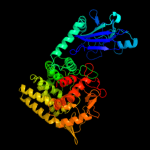 |
99.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucoamylase;
PDBTitle: crystal structure of bacterial glucoamylase
|
| 11 | c3cihA_
|
|
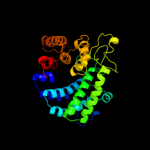 |
99.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alpha-rhamnosidase;
PDBTitle: crystal structure of a putative alpha-rhamnosidase from2 bacteroides thetaiotaomicron
|
| 12 | d1ulva1
|
|
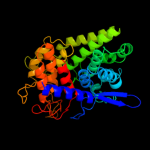 |
99.8 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Bacterial glucoamylase C-terminal domain-like |
| 13 | d1gaia_
|
|
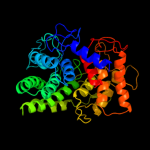 |
99.7 |
18 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glucoamylase |
| 14 | c1ug9A_
|
|
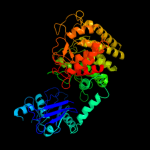 |
99.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucodextranase;
PDBTitle: crystal structure of glucodextranase from arthrobacter globiformis i42
|
| 15 | d2fbaa1
|
|
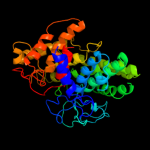 |
99.7 |
17 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glucoamylase |
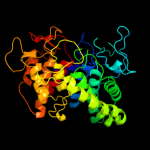
| 16 | c2vn4A_
|
|
 |
99.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucoamylase;
PDBTitle: glycoside hydrolase family 15 glucoamylase from hypocrea2 jecorina
|
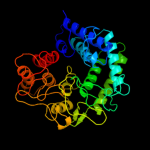
| 17 | c3qspB_
|
|
 |
98.9 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: analysis of a new family of widely distributed metal-independent alpha2 mannosidases provides unique insight into the processing of n-linked3 glycans, streptococcus pneumoniae sp_2144 non-productive substrate4 complex with alpha-1,6-mannobiose
|
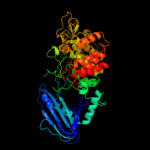
| 18 | c2eacB_
|
|
 |
98.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-fucosidase;
PDBTitle: crystal structure of 1,2-a-l-fucosidase from2 bifidobacterium bifidum in complex with3 deoxyfuconojirimycin
|
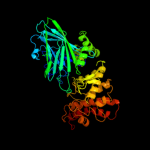
| 19 | c2ww1B_
|
|
 |
98.7 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-1,2-mannosidase;
PDBTitle: structure of the family gh92 inverting mannosidase bt39902 from bacteroides thetaiotaomicron vpi-5482 in complex with3 thiomannobioside
|
| 20 | d2nvpa1
|
|
 |
98.7 |
15 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:CPF0428-like |
| 21 | d1h54a1 |
|
not modelled |
98.7 |
13 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glycosyltransferase family 36 C-terminal domain |
| 22 | c2wvyA_ |
|
not modelled |
98.6 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-1,2-mannosidase;
PDBTitle: structure of the family gh92 inverting mannosidase bt21992 from bacteroides thetaiotaomicron vpi-5482
|
| 23 | c2p0vA_ |
|
not modelled |
98.6 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein bt3781;
PDBTitle: crystal structure of bt3781 protein from bacteroides2 thetaiotaomicron, northeast structural genomics target3 btr58
|
| 24 | d2p0va1 |
|
not modelled |
98.6 |
19 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:CPF0428-like |
| 25 | c1h54B_ |
|
not modelled |
98.2 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:maltose phosphorylase;
PDBTitle: maltose phosphorylase from lactobacillus brevis
|
| 26 | c2rdyB_ |
|
not modelled |
98.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:bh0842 protein;
PDBTitle: crystal structure of a putative glycoside hydrolase family2 protein from bacillus halodurans
|
| 27 | c3gt5A_ |
|
not modelled |
96.3 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylglucosamine 2-epimerase;
PDBTitle: crystal structure of an n-acetylglucosamine 2-epimerase family protein2 from xylella fastidiosa
|
| 28 | d2afaa1 |
|
not modelled |
95.1 |
15 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:N-acylglucosamine (NAG) epimerase |
| 29 | d1fp3a_ |
|
not modelled |
94.1 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:N-acylglucosamine (NAG) epimerase |
| 30 | d2d5ja1 |
|
not modelled |
91.1 |
16 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glycosyl Hydrolase Family 88 |
| 31 | c2zzrA_ |
|
not modelled |
87.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:unsaturated glucuronyl hydrolase;
PDBTitle: crystal structure of unsaturated glucuronyl hydrolase from2 streptcoccus agalactiae
|
| 32 | c1ut9A_ |
|
not modelled |
70.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellulose 1,4-beta-cellobiosidase;
PDBTitle: structural basis for the exocellulase activity of the2 cellobiohydrolase cbha from c. thermocellum
|
| 33 | c3k11A_ |
|
not modelled |
70.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative glycosyl hydrolase;
PDBTitle: crystal structure of putative glycosyl hydrolase (np_813087.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.80 a resolution
|
| 34 | d1ut9a1 |
|
not modelled |
65.6 |
14 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Cellulases catalytic domain |
| 35 | d1nc5a_ |
|
not modelled |
62.2 |
11 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Hypothetical protein YteR |
| 36 | c1ga2A_ |
|
not modelled |
61.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase 9g;
PDBTitle: the crystal structure of endoglucanase 9g from clostridium2 cellulolyticum complexed with cellobiose
|
| 37 | d1ia6a_ |
|
not modelled |
60.6 |
14 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Cellulases catalytic domain |
| 38 | c1clcA_ |
|
not modelled |
55.5 |
15 |
PDB header:glycosyl hydrolase
Chain: A: PDB Molecule:endoglucanase celd; ec: 3.2.1.4;
PDBTitle: three-dimensional structure of endoglucanase d at 1.92 angstroms resolution
|
| 39 | c3pmmA_ |
|
not modelled |
54.7 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative cytoplasmic protein;
PDBTitle: the crystal structure of a possible member of gh105 family from2 klebsiella pneumoniae subsp. pneumoniae mgh 78578
|
| 40 | d1clca1 |
|
not modelled |
52.7 |
12 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Cellulases catalytic domain |
| 41 | d1g87a1 |
|
not modelled |
50.1 |
19 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Cellulases catalytic domain |
| 42 | c2xfgA_ |
|
not modelled |
43.6 |
14 |
PDB header:hydrolase/sugar binding protein
Chain: A: PDB Molecule:endoglucanase 1;
PDBTitle: reassembly and co-crystallization of a family 9 processive2 endoglucanase from separately expressed gh9 and cbm3c3 modules
|
| 43 | c3gzkA_ |
|
not modelled |
40.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellulase;
PDBTitle: structure of a. acidocaldarius cellulase cela
|
| 44 | c3nrdB_ |
|
not modelled |
30.5 |
22 |
PDB header:nucleotide binding protein
Chain: B: PDB Molecule:histidine triad (hit) protein;
PDBTitle: crystal structure of a histidine triad (hit) protein (smc02904) from2 sinorhizobium meliloti 1021 at 2.06 a resolution
|
| 45 | d1tf4a1 |
|
not modelled |
29.2 |
12 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Cellulases catalytic domain |
| 46 | d1ks8a_ |
|
not modelled |
27.4 |
10 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Cellulases catalytic domain |
| 47 | c1x9dA_ |
|
not modelled |
25.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoplasmic reticulum mannosyl-oligosaccharide 1,
PDBTitle: crystal structure of human class i alpha-1,2-mannosidase in2 complex with thio-disaccharide substrate analogue
|
| 48 | d1x9da1 |
|
not modelled |
25.3 |
17 |
Fold:alpha/alpha toroid
Superfamily:Seven-hairpin glycosidases
Family:Class I alpha-1;2-mannosidase, catalytic domain |
| 49 | d1of5b_ |
|
not modelled |
22.0 |
19 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:NTF2-like |
| 50 | d2h8pc1 |
|
not modelled |
17.6 |
7 |
Fold:Voltage-gated potassium channels
Superfamily:Voltage-gated potassium channels
Family:Voltage-gated potassium channels |
| 51 | d2pifa1 |
|
not modelled |
16.3 |
15 |
Fold:PSTPO5379-like
Superfamily:PSTPO5379-like
Family:PSTPO5379-like |
| 52 | c3oheA_ |
|
not modelled |
16.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:histidine triad (hit) protein;
PDBTitle: crystal structure of a histidine triad protein (maqu_1709) from2 marinobacter aquaeolei vt8 at 1.20 a resolution
|
| 53 | c3k7xA_ |
|
not modelled |
15.4 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin0763 protein;
PDBTitle: crystal structure of the lin0763 protein from listeria2 innocua. northeast structural genomics consortium target3 lkr23.
|
| 54 | c2lbbA_ |
|
not modelled |
12.7 |
27 |
PDB header:protein binding
Chain: A: PDB Molecule:acyl coa binding protein;
PDBTitle: solution structure of acyl coa binding protein from babesia bovis t2bo
|
| 55 | c1xc0A_ |
|
not modelled |
12.4 |
41 |
PDB header:signaling protein
Chain: A: PDB Molecule:pardaxin p-4;
PDBTitle: twenty lowest energy structures of pa4 by solution nmr
|
| 56 | d1nzpa_ |
|
not modelled |
12.3 |
15 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 57 | c3sohB_ |
|
not modelled |
11.2 |
20 |
PDB header:motor protein
Chain: B: PDB Molecule:flagellar motor switch protein flig;
PDBTitle: architecture of the flagellar rotor
|
| 58 | d1i39a_ |
|
not modelled |
11.1 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 59 | c1i3aA_ |
|
not modelled |
11.1 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hii;
PDBTitle: rnase hii from archaeoglobus fulgidus with cobalt hexammine2 chloride
|
| 60 | d1gwya_ |
|
not modelled |
10.9 |
11 |
Fold:Cytolysin/lectin
Superfamily:Cytolysin/lectin
Family:Anemone pore-forming cytolysin |
| 61 | c2xtdB_ |
|
not modelled |
10.6 |
8 |
PDB header:transcription
Chain: B: PDB Molecule:tbl1 f-box-like/wd repeat-containing protein tbl1x;
PDBTitle: structure of the tbl1 tetramerisation domain
|
| 62 | c3db3A_ |
|
not modelled |
10.2 |
43 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase uhrf1;
PDBTitle: crystal structure of the tandem tudor domains of the e3 ubiquitin-2 protein ligase uhrf1 in complex with trimethylated histone h3-k93 peptide
|
| 63 | c2xteH_ |
|
not modelled |
9.8 |
8 |
PDB header:transcription
Chain: H: PDB Molecule:f-box-like/wd repeat-containing protein tbl1x;
PDBTitle: structure of the tbl1 tetramerisation domain
|
| 64 | c2r7aC_ |
|
not modelled |
9.5 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:bacterial heme binding protein;
PDBTitle: crystal structure of a periplasmic heme binding protein2 from shigella dysenteriae
|
| 65 | c3kioA_ |
|
not modelled |
9.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease h2 subunit a;
PDBTitle: mouse rnase h2 complex
|
| 66 | c2axtc_ |
|
not modelled |
9.0 |
16 |
PDB header:electron transport
Chain: C: PDB Molecule:photosystem ii cp43 protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
|
| 67 | d2axtc1 |
|
not modelled |
9.0 |
16 |
Fold:Photosystem II antenna protein-like
Superfamily:Photosystem II antenna protein-like
Family:Photosystem II antenna protein-like |
| 68 | d1iwpa_ |
|
not modelled |
8.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Diol dehydratase, alpha subunit |
| 69 | d1jmsa1 |
|
not modelled |
8.9 |
10 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 70 | c2jw1A_ |
|
not modelled |
8.8 |
21 |
PDB header:membrane protein
Chain: A: PDB Molecule:lipoprotein mxim;
PDBTitle: structural characterization of the type iii pilotin-2 secretin interaction in shigella flexneri by nmr3 spectroscopy
|
| 71 | c3i4sB_ |
|
not modelled |
8.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidine triad protein;
PDBTitle: crystal structure of histidine triad protein blr8122 from2 bradyrhizobium japonicum
|
| 72 | d3boja1 |
|
not modelled |
8.6 |
20 |
Fold:CdCA1 repeat-like
Superfamily:CdCA1 repeat-like
Family:CdCA1 repeat-like |
| 73 | d1ekea_ |
|
not modelled |
8.5 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 74 | c2qx5B_ |
|
not modelled |
8.4 |
21 |
PDB header:transport protein
Chain: B: PDB Molecule:nucleoporin nic96;
PDBTitle: structure of nucleoporin nic96
|
| 75 | d1uaxa_ |
|
not modelled |
8.4 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 76 | c2vixB_ |
|
not modelled |
8.3 |
14 |
PDB header:transport protein
Chain: B: PDB Molecule:protein mxic;
PDBTitle: methylated shigella flexneri mxic
|
| 77 | c2d0bA_ |
|
not modelled |
8.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hiii;
PDBTitle: crystal structure of bst-rnase hiii in complex with mg2+
|
| 78 | d3boea1 |
|
not modelled |
8.2 |
23 |
Fold:CdCA1 repeat-like
Superfamily:CdCA1 repeat-like
Family:CdCA1 repeat-like |
| 79 | c3i24B_ |
|
not modelled |
8.1 |
43 |
PDB header:hydrolase
Chain: B: PDB Molecule:hit family hydrolase;
PDBTitle: crystal structure of a hit family hydrolase protein from2 vibrio fischeri. northeast structural genomics consortium3 target id vfr176
|
| 80 | d1w8ia_ |
|
not modelled |
8.0 |
16 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:PIN domain |
| 81 | d2pbea2 |
|
not modelled |
7.9 |
26 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:AadK N-terminal domain-like |
| 82 | c2r2vB_ |
|
not modelled |
7.6 |
36 |
PDB header:de novo protein
Chain: B: PDB Molecule:gcn4 leucine zipper;
PDBTitle: sequence determinants of the topology of the lac repressor2 tetrameric coiled coil
|
| 83 | d1io2a_ |
|
not modelled |
7.6 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 84 | c1ichA_ |
|
not modelled |
7.3 |
0 |
PDB header:apoptosis
Chain: A: PDB Molecule:tumor necrosis factor receptor-1;
PDBTitle: solution structure of the tumor necrosis factor receptor-12 death domain
|
| 85 | d1icha_ |
|
not modelled |
7.3 |
0 |
Fold:DEATH domain
Superfamily:DEATH domain
Family:DEATH domain, DD |
| 86 | d1h2vc2 |
|
not modelled |
7.2 |
14 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:MIF4G domain-like |
| 87 | c3igmA_ |
|
not modelled |
7.0 |
16 |
PDB header:transcription/dna
Chain: A: PDB Molecule:pf14_0633 protein;
PDBTitle: a 2.2a crystal structure of the ap2 domain of pf14_0633 from p.2 falciparum, bound as a domain-swapped dimer to its cognate dna
|
| 88 | d2bcqa1 |
|
not modelled |
7.0 |
17 |
Fold:SAM domain-like
Superfamily:DNA polymerase beta, N-terminal domain-like
Family:DNA polymerase beta, N-terminal domain-like |
| 89 | c3fgrB_ |
|
not modelled |
6.8 |
32 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative phospholipase b-like 2 40 kda form;
PDBTitle: two chain form of the 66.3 kda protein at 1.8 angstroem
|
| 90 | c2fqzC_ |
|
not modelled |
6.7 |
13 |
PDB header:hydrolase/dna
Chain: C: PDB Molecule:r.ecl18ki;
PDBTitle: metal-depleted ecl18ki in complex with uncleaved dna
|
| 91 | c3hwjA_ |
|
not modelled |
6.7 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase mycbp2;
PDBTitle: crystal structure of the second phr domain of mouse myc-2 binding protein 2 (mycbp-2)
|
| 92 | d2d5ua1 |
|
not modelled |
6.6 |
10 |
Fold:PUG domain-like
Superfamily:PUG domain-like
Family:PUG domain |
| 93 | c2bldD_ |
|
not modelled |
6.5 |
17 |
PDB header:virus
Chain: D: PDB Molecule:penton protein;
PDBTitle: the quasi-atomic model of human adenovirus type 52 capsid (part 1)
|
| 94 | c3renB_ |
|
not modelled |
6.5 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase, family 8;
PDBTitle: cpf_2247, a novel alpha-amylase from clostridium perfringens
|
| 95 | d1su3a1 |
|
not modelled |
6.4 |
17 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
| 96 | d2etja1 |
|
not modelled |
6.4 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 97 | c2etjA_ |
|
not modelled |
6.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hii;
PDBTitle: crystal structure of ribonuclease hii (ec 3.1.26.4) (rnase hii)2 (tm0915) from thermotoga maritima at 1.74 a resolution
|
| 98 | d3pmga4 |
|
not modelled |
6.3 |
20 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 99 | c3no8A_ |
|
not modelled |
6.3 |
25 |
PDB header:isomerase regulator
Chain: A: PDB Molecule:btb/poz domain-containing protein 2;
PDBTitle: crystal structure of the phr domain from human btbd2 protein
|