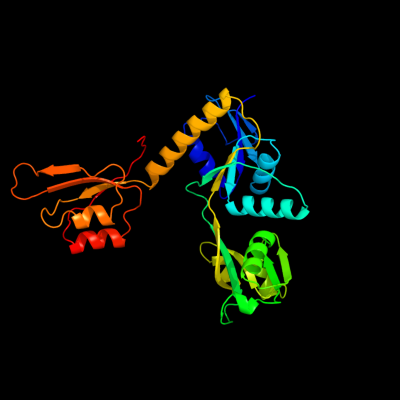
| 1 | c1q1kA_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: structure of atp-phosphoribosyltransferase from e. coli complexed with2 pr-atp
|
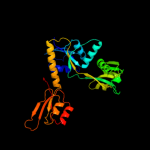
| 2 | c2vd3B_
|
|
 |
100.0 |
37 |
PDB header:transferase
Chain: B: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: the structure of histidine inhibited hisg from2 methanobacterium thermoautotrophicum
|
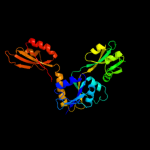
| 3 | c1nh7A_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: atp phosphoribosyltransferase (atp-prtase) from mycobacterium2 tuberculosis
|
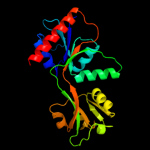
| 4 | d1h3da1
|
|
 |
100.0 |
100 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
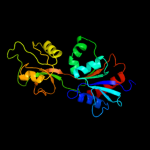
| 5 | c2vd2A_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: the crystal structure of hisg from b. subtilis
|
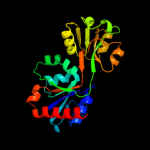
| 6 | d1nh8a1
|
|
 |
100.0 |
31 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
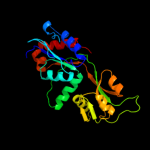
| 7 | d1ve4a1
|
|
 |
100.0 |
34 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 8 | d1o63a_
|
|
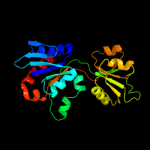 |
100.0 |
30 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 9 | d1z7me1
|
|
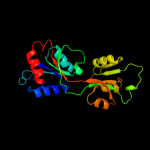 |
100.0 |
32 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 10 | d1nh8a2
|
|
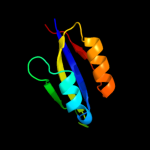 |
99.9 |
33 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain |
| 11 | d1h3da2
|
|
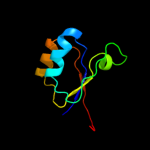 |
99.9 |
100 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain |
| 12 | c3uifA_
|
|
 |
98.0 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:sulfonate abc transporter, periplasmic sulfonate-binding
PDBTitle: crystal structure of putative sulfonate abc transporter, periplasmic2 sulfonate-binding protein ssua from methylobacillus flagellatus kt
|
| 13 | c3ix1A_
|
|
 |
98.0 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding
PDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
|
| 14 | c3ix1B_
|
|
 |
98.0 |
17 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding
PDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
|
| 15 | c3qslA_
|
|
 |
97.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: structure of cae31940 from bordetella bronchiseptica rb50
|
| 16 | c3e4rA_
|
|
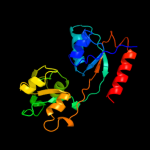 |
97.5 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein;
PDBTitle: crystal structure of the alkanesulfonate binding protein2 (ssua) from the phytopathogenic bacteria xanthomonas3 axonopodis pv. citri bound to hepes
|
| 17 | c2x26A_
|
|
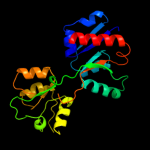 |
97.4 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic aliphatic sulphonates-binding protein;
PDBTitle: crystal structure of the periplasmic aliphatic sulphonate2 binding protein ssua from escherichia coli
|
| 18 | c3hn0A_
|
|
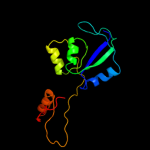 |
97.3 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein;
PDBTitle: crystal structure of an abc transporter (bdi_1369) from2 parabacteroides distasonis at 1.75 a resolution
|
| 19 | c3un6A_
|
|
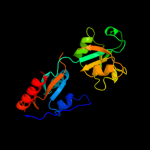 |
96.8 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein saouhsc_00137;
PDBTitle: 2.0 angstrom crystal structure of ligand binding component of abc-type2 import system from staphylococcus aureus with zinc bound
|
| 20 | d2nxoa1
|
|
 |
96.7 |
15 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 21 | c2de4B_ |
|
not modelled |
96.6 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:dibenzothiophene desulfurization enzyme b;
PDBTitle: crystal structure of dszb c27s mutant in complex with biphenyl-2-2 sulfinic acid
|
| 22 | c2x7pA_ |
|
not modelled |
96.4 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:possible thiamine biosynthesis enzyme;
PDBTitle: the conserved candida albicans ca3427 gene product defines a new2 family of proteins exhibiting the generic periplasmic binding3 protein structural fold
|
| 23 | c1p99A_ |
|
not modelled |
96.2 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pg110;
PDBTitle: 1.7a crystal structure of protein pg110 from staphylococcus2 aureus
|
| 24 | d1p99a_ |
|
not modelled |
96.2 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 25 | d2czla1 |
|
not modelled |
95.8 |
17 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 26 | c3n5lA_ |
|
not modelled |
95.3 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:binding protein component of abc phosphonate transporter;
PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
|
| 27 | d1zbma1 |
|
not modelled |
95.1 |
16 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 28 | c2pfzA_ |
|
not modelled |
95.0 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of dctp6, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
|
| 29 | c3fxbB_ |
|
not modelled |
94.4 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:trap dicarboxylate transporter, dctp subunit;
PDBTitle: crystal structure of the ectoine-binding protein ueha
|
| 30 | c3tmgA_ |
|
not modelled |
94.4 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine, l-proline abc transporter,
PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
|
| 31 | c3b50A_ |
|
not modelled |
92.0 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:sialic acid-binding periplasmic protein siap;
PDBTitle: structure of h. influenzae sialic acid binding protein2 bound to neu5ac.
|
| 32 | c3l6gA_ |
|
not modelled |
91.2 |
9 |
PDB header:glycine betaine-binding protein
Chain: A: PDB Molecule:betaine abc transporter permease and substrate binding
PDBTitle: crystal structure of lactococcal opuac in its open conformation
|
| 33 | c3kzgB_ |
|
not modelled |
90.4 |
15 |
PDB header:transport protein
Chain: B: PDB Molecule:arginine 3rd transport system periplasmic binding
PDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
|
| 34 | c2hpgB_ |
|
not modelled |
89.4 |
17 |
PDB header:ligand binding protein
Chain: B: PDB Molecule:abc transporter, periplasmic substrate-binding
PDBTitle: the crystal structure of a thermophilic trap periplasmic2 binding protein
|
| 35 | c2zzxD_ |
|
not modelled |
88.9 |
13 |
PDB header:transport protein
Chain: D: PDB Molecule:abc transporter, solute-binding protein;
PDBTitle: crystal structure of a periplasmic substrate binding protein in2 complex with lactate
|
| 36 | c2hzkB_ |
|
not modelled |
88.5 |
13 |
PDB header:ligand binding, transport protein
Chain: B: PDB Molecule:trap-t family sorbitol/mannitol transporter, periplasmic
PDBTitle: crystal structures of a sodium-alpha-keto acid binding subunit from a2 trap transporter in its open form
|
| 37 | c2rd5D_ |
|
not modelled |
88.1 |
11 |
PDB header:protein binding
Chain: D: PDB Molecule:pii protein;
PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
|
| 38 | c2pfyA_ |
|
not modelled |
88.0 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of dctp7, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
|
| 39 | c3ir1F_ |
|
not modelled |
87.5 |
14 |
PDB header:protein binding
Chain: F: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
|
| 40 | c2g29A_ |
|
not modelled |
87.0 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein nrta;
PDBTitle: crystal structure of the periplasmic nitrate-binding2 protein nrta from synechocystis pcc 6803
|
| 41 | c3gxaA_ |
|
not modelled |
86.0 |
9 |
PDB header:protein binding
Chain: A: PDB Molecule:outer membrane lipoprotein gna1946;
PDBTitle: crystal structure of gna1946
|
| 42 | d1vm0a_ |
|
not modelled |
82.7 |
34 |
Fold:IF3-like
Superfamily:AlbA-like
Family:Hypothetical protein At2g34160 |
| 43 | d1us5a_ |
|
not modelled |
81.5 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 44 | c3l7pA_ |
|
not modelled |
80.4 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:putative nitrogen regulatory protein pii;
PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
|
| 45 | c2q3vB_ |
|
not modelled |
80.3 |
34 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein at2g34160;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at2g34160
|
| 46 | c2vpnB_ |
|
not modelled |
79.2 |
17 |
PDB header:transport
Chain: B: PDB Molecule:periplasmic substrate binding protein;
PDBTitle: high-resolution structure of the periplasmic ectoine-2 binding protein from teaabc trap-transporter of halomonas3 elongata
|
| 47 | c3gyyC_ |
|
not modelled |
77.6 |
17 |
PDB header:transport protein
Chain: C: PDB Molecule:periplasmic substrate binding protein;
PDBTitle: the ectoine binding protein of the teaabc trap transporter teaa in the2 apo-state
|
| 48 | c3bzqA_ |
|
not modelled |
75.7 |
9 |
PDB header:signaling protein/transcription
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii;
PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
|
| 49 | d1xs5a_ |
|
not modelled |
73.9 |
15 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 50 | c2j9dG_ |
|
not modelled |
73.8 |
8 |
PDB header:membrane transport
Chain: G: PDB Molecule:hypothetical nitrogen regulatory pii-like
PDBTitle: structure of glnk1 with bound effectors indicates2 regulatory mechanism for ammonia uptake
|
| 51 | d1qy7a_ |
|
not modelled |
73.7 |
12 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 52 | c3mhyC_ |
|
not modelled |
72.5 |
18 |
PDB header:signaling protein
Chain: C: PDB Molecule:pii-like protein pz;
PDBTitle: a new pii protein structure
|
| 53 | c3tqwA_ |
|
not modelled |
72.3 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:methionine-binding protein;
PDBTitle: structure of a abc transporter, periplasmic substrate-binding protein2 from coxiella burnetii
|
| 54 | d2ns1b1 |
|
not modelled |
72.3 |
12 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 55 | d2piia_ |
|
not modelled |
68.8 |
12 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 56 | c3ho7A_ |
|
not modelled |
68.2 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:oxyr;
PDBTitle: crystal structure of oxyr from porphyromonas gingivalis
|
| 57 | c3o8wA_ |
|
not modelled |
67.8 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii (glnb-1);
PDBTitle: archaeoglobus fulgidus glnk1
|
| 58 | d1ul3a_ |
|
not modelled |
65.8 |
12 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 59 | c2hxrA_ |
|
not modelled |
65.6 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional regulator cynr;
PDBTitle: structure of the ligand binding domain of e. coli cynr, a2 transcriptional regulator controlling cyanate metabolism
|
| 60 | d1hwua_ |
|
not modelled |
64.9 |
15 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 61 | d1eh3a_ |
|
not modelled |
62.1 |
8 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 62 | c2f5xC_ |
|
not modelled |
61.7 |
11 |
PDB header:transport protein
Chain: C: PDB Molecule:bugd;
PDBTitle: structure of periplasmic binding protein bugd
|
| 63 | c2i4cA_ |
|
not modelled |
59.1 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:bicarbonate transporter;
PDBTitle: crystal structure of bicarbonate transport protein cmpa from2 synechocystis sp. pcc 6803 in complex with bicarbonate and calcium
|
| 64 | c2rc9A_ |
|
not modelled |
58.3 |
12 |
PDB header:membrane protein
Chain: A: PDB Molecule:glutamate [nmda] receptor subunit 3a;
PDBTitle: crystal structure of the nr3a ligand binding core complex with acpc at2 1.96 angstrom resolution
|
| 65 | c3c7bA_ |
|
not modelled |
55.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit alpha;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 66 | c3gmgB_ |
|
not modelled |
54.2 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein rv1825/mt1873;
PDBTitle: crystal structure of an uncharacterized conserved protein2 from mycobacterium tuberculosis
|
| 67 | d1dtza1 |
|
not modelled |
53.5 |
6 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 68 | d1i6aa_ |
|
not modelled |
52.4 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 69 | d3c7bb2 |
|
not modelled |
47.8 |
13 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |
| 70 | d2cz4a1 |
|
not modelled |
46.8 |
24 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 71 | c3ncpD_ |
|
not modelled |
43.7 |
25 |
PDB header:signaling protein
Chain: D: PDB Molecule:nitrogen regulatory protein p-ii (glnb-2);
PDBTitle: glnk2 from archaeoglobus fulgidus
|
| 72 | c3ce8A_ |
|
not modelled |
43.1 |
10 |
PDB header:unknown function
Chain: A: PDB Molecule:putative pii-like nitrogen regulatory protein;
PDBTitle: crystal structure of a duf3240 family protein (sbal_0098) from2 shewanella baltica os155 at 2.40 a resolution
|
| 73 | d1ryoa_ |
|
not modelled |
41.8 |
10 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 74 | d1ce2a1 |
|
not modelled |
41.4 |
7 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 75 | d2dy1a5 |
|
not modelled |
39.6 |
17 |
Fold:Ferredoxin-like
Superfamily:EF-G C-terminal domain-like
Family:EF-G/eEF-2 domains III and V |
| 76 | c3mstA_ |
|
not modelled |
39.5 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:putative nitrate transport protein;
PDBTitle: crystal structure of a putative nitrate transport protein (tvn0104)2 from thermoplasma volcanium at 1.35 a resolution
|
| 77 | c3k2dA_ |
|
not modelled |
37.9 |
18 |
PDB header:immune system
Chain: A: PDB Molecule:abc-type metal ion transport system, periplasmic component;
PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
|
| 78 | c2dvzA_ |
|
not modelled |
37.7 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: structure of a periplasmic transporter
|
| 79 | c2ylnA_ |
|
not modelled |
36.6 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter, periplasmic binding protein,
PDBTitle: crystal structure of the l-cystine solute receptor of2 neisseria gonorrhoeae in the closed conformation
|
| 80 | c3onmB_ |
|
not modelled |
36.3 |
18 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator lrha;
PDBTitle: effector binding domain of lysr-type transcription factor rovm from y.2 pseudotuberculosis
|
| 81 | d1h76a1 |
|
not modelled |
36.0 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 82 | d1pb7a_ |
|
not modelled |
35.4 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 83 | c2qpqC_ |
|
not modelled |
35.0 |
7 |
PDB header:transport protein
Chain: C: PDB Molecule:protein bug27;
PDBTitle: structure of bug27 from bordetella pertussis
|
| 84 | d1ieja_ |
|
not modelled |
32.9 |
10 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 85 | c3kbrA_ |
|
not modelled |
31.4 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:cyclohexadienyl dehydratase;
PDBTitle: the crystal structure of cyclohexadienyl dehydratase precursor from2 pseudomonas aeruginosa pa01
|
| 86 | c2pyyB_ |
|
not modelled |
29.9 |
19 |
PDB header:transport protein
Chain: B: PDB Molecule:ionotropic glutamate receptor bacterial homologue;
PDBTitle: crystal structure of the glur0 ligand-binding core from nostoc2 punctiforme in complex with (l)-glutamate
|
| 87 | d1ufra_ |
|
not modelled |
29.8 |
45 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 88 | d1w30a_ |
|
not modelled |
29.3 |
18 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 89 | d1b1xa1 |
|
not modelled |
29.0 |
10 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 90 | c3i31A_ |
|
not modelled |
27.6 |
13 |
PDB header:rna binding protein,hydrolase
Chain: A: PDB Molecule:heat resistant rna dependent atpase;
PDBTitle: hera helicase rna binding domain is an rrm fold
|
| 91 | d2hava1 |
|
not modelled |
27.2 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 92 | d1a3ca_ |
|
not modelled |
25.8 |
45 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 93 | d1vcha1 |
|
not modelled |
25.7 |
45 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 94 | d1cjba_ |
|
not modelled |
25.6 |
45 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 95 | d2fyia1 |
|
not modelled |
25.5 |
19 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 96 | d1g9sa_ |
|
not modelled |
25.2 |
36 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 97 | d1jw1a1 |
|
not modelled |
24.9 |
8 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Transferrin |
| 98 | d2igba1 |
|
not modelled |
24.0 |
45 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 99 | d1tc1a_ |
|
not modelled |
23.7 |
45 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 100 | d1vfja_ |
|
not modelled |
23.3 |
27 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 101 | c3kb8A_ |
|
not modelled |
23.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine phosphoribosyltransferase;
PDBTitle: 2.09 angstrom resolution structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-1) from bacillus anthracis str. 'ames3 ancestor' in complex with gmp
|
| 102 | d1z7ga1 |
|
not modelled |
22.9 |
36 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 103 | d1e0ta3 |
|
not modelled |
22.8 |
27 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
| 104 | d1fsga_ |
|
not modelled |
22.6 |
55 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 105 | c3m70A_ |
|
not modelled |
22.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein tehb homolog;
PDBTitle: crystal structure of tehb from haemophilus influenzae
|
| 106 | d1nula_ |
|
not modelled |
22.5 |
64 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 107 | d1hgxa_ |
|
not modelled |
22.3 |
36 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 108 | d1p17b_ |
|
not modelled |
21.9 |
45 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 109 | c3o7mD_ |
|
not modelled |
21.8 |
36 |
PDB header:transferase
Chain: D: PDB Molecule:hypoxanthine phosphoribosyltransferase;
PDBTitle: 1.98 angstrom resolution crystal structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-2) from bacillus anthracis str. 'ames3 ancestor'
|
| 110 | c2jbhA_ |
|
not modelled |
21.8 |
55 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosyltransferase domain-containing protein 1;
PDBTitle: human phosphoribosyl transferase domain containing 1
|
| 111 | d2g50a3 |
|
not modelled |
21.8 |
35 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
| 112 | c2o1mB_ |
|
not modelled |
21.8 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:probable amino-acid abc transporter
PDBTitle: crystal structure of the probable amino-acid abc2 transporter extracellular-binding protein ytmk from3 bacillus subtilis. northeast structural genomics4 consortium target sr572
|
| 113 | d1g2qa_ |
|
not modelled |
21.7 |
36 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 114 | d1ii5a_ |
|
not modelled |
21.6 |
11 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 115 | d1vdma1 |
|
not modelled |
20.5 |
43 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 116 | c2ywtA_ |
|
not modelled |
20.5 |
55 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: crystal structure of hypoxanthine-guanine2 phosphoribosyltransferase with gmp from thermus3 thermophilus hb8
|
| 117 | d1gnla_ |
|
not modelled |
20.2 |
17 |
Fold:Prismane protein-like
Superfamily:Prismane protein-like
Family:Hybrid cluster protein (prismane protein) |