1 c2zxrA_
100.0
35
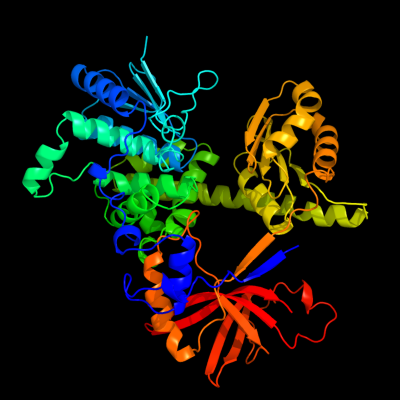
PDB header: hydrolaseChain: A: PDB Molecule: single-stranded dna specific exonuclease recj;PDBTitle: crystal structure of recj in complex with mg2+ from thermus2 thermophilus hb8
2 d1ir6a_
100.0
38
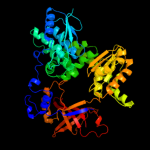
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Exonuclease RecJ3 c1ir6A_
100.0
38
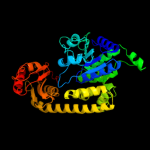
PDB header: hydrolaseChain: A: PDB Molecule: exonuclease recj;PDBTitle: crystal structure of exonuclease recj bound to manganese
4 c3dmaA_
100.0
16
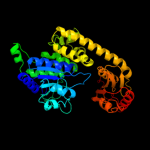
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: exopolyphosphatase-related protein;PDBTitle: crystal structure of an exopolyphosphatase-related protein2 from bacteroides fragilis. northeast structural genomics3 target bfr192
5 c3devB_
100.0
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: sh1221;PDBTitle: crystal structure of sh1221 protein from staphylococcus haemolyticus,2 northeast structural genomics consortium target shr87
6 d1wpna_
98.0
13
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)7 d1i74a_
98.0
19
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)8 d1k20a_
97.7
15
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)9 d2hawa1
97.6
11
Fold: DHH phosphoesterasesSuperfamily: DHH phosphoesterasesFamily: Manganese-dependent inorganic pyrophosphatase (family II)10 c2eb0B_
97.3
16
PDB header: hydrolaseChain: B: PDB Molecule: manganese-dependent inorganic pyrophosphatase;PDBTitle: crystal structure of methanococcus jannaschii putative family ii2 inorganic pyrophosphatase
11 c2zvfG_
95.8
14
PDB header: ligaseChain: G: PDB Molecule: alanyl-trna synthetase;PDBTitle: crystal structure of archaeoglobus fulgidus alanyl-trna2 synthetase c-terminal dimerization domain
12 c2qb6A_
94.3
18
PDB header: hydrolaseChain: A: PDB Molecule: exopolyphosphatase;PDBTitle: saccharomyces cerevisiae cytosolic exopolyphosphatase, sulfate complex
13 d2fywa1
91.9
19
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like14 d1x94a_
91.7
14
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain15 c1nriA_
91.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein hi0754;PDBTitle: crystal structure of putative phosphosugar isomerase hi0754 from2 haemophilus influenzae
16 d1nria_
91.3
20
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain17 c2yvaB_
89.8
14
PDB header: dna binding proteinChain: B: PDB Molecule: dnaa initiator-associating protein diaa;PDBTitle: crystal structure of escherichia coli diaa
18 d1nmpa_
89.6
21
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like19 d1qo2a_
88.2
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes20 d2gx8a1
85.4
19
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like21 d1h5ya_
not modelled
85.2
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes22 c3g98B_
not modelled
85.2
14
PDB header: ligaseChain: B: PDB Molecule: alanyl-trna synthetase;PDBTitle: crystal structure of the c-ala domain from aquifex aeolicus2 alanyl-trna synthetase
23 c2nydB_
not modelled
84.4
20
PDB header: unknown functionChain: B: PDB Molecule: upf0135 protein sa1388;PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
24 c2ihfA_
not modelled
84.0
11
PDB header: dna binding proteinChain: A: PDB Molecule: single-stranded dna-binding protein;PDBTitle: crystal structure of deletion mutant delta 228-252 r190a of the2 single-stranded dna binding protein from thermus aquaticus
25 d2jfga1
not modelled
83.4
18
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain26 c3trjC_
not modelled
83.3
16
PDB header: isomeraseChain: C: PDB Molecule: phosphoheptose isomerase;PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
27 d1tk9a_
not modelled
82.1
16
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain28 c1ecjB_
not modelled
81.8
19
PDB header: transferaseChain: B: PDB Molecule: glutamine phosphoribosylpyrophosphatePDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
29 c2gx8B_
not modelled
81.2
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal stucture of bacillus cereus protein related to nif3
30 c2w6rA_
not modelled
80.8
13
PDB header: lyaseChain: A: PDB Molecule: imidazole glycerol phosphate synthase subunitPDBTitle: crystal structure of an artificial (ba)8-barrel protein2 designed from identical half barrels
31 c3cvjB_
not modelled
79.0
18
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
32 d1vzwa1
not modelled
78.5
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes33 c3pdiG_
not modelled
78.4
11
PDB header: protein bindingChain: G: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nife;PDBTitle: precursor bound nifen
34 c3kf6A_
not modelled
78.1
20
PDB header: structural proteinChain: A: PDB Molecule: protein stn1;PDBTitle: crystal structure of s. pombe stn1-ten1 complex
35 c2iheA_
not modelled
78.0
9
PDB header: dna binding proteinChain: A: PDB Molecule: single-stranded dna-binding protein;PDBTitle: crystal structure of wild-type single-stranded dna binding protein2 from thermus aquaticus
36 d1y5ia2
not modelled
77.9
14
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-337 d1p3da1
not modelled
77.5
21
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain38 c3q98A_
not modelled
77.4
22
PDB header: transferaseChain: A: PDB Molecule: transcarbamylase;PDBTitle: structure of ygew encoded protein from e. coli
39 d1m3sa_
not modelled
77.4
15
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain40 c3ippA_
not modelled
76.5
19
PDB header: transferaseChain: A: PDB Molecule: putative thiosulfate sulfurtransferase ynje;PDBTitle: crystal structure of sulfur-free ynje
41 c1zq2A_
not modelled
76.3
18
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: crystal structure of n-acetyl-l-ornithine transcarbamylase2 complexed with cp
42 d1x92a_
not modelled
76.3
13
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain43 d1u9ya1
not modelled
75.9
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like44 d1pvda1
not modelled
74.4
14
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain45 c2p4dA_
not modelled
74.0
10
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase;PDBTitle: structure-assisted discovery of variola major h12 phosphatase inhibitors
46 c3tdmD_
not modelled
73.8
15
PDB header: de novo proteinChain: D: PDB Molecule: computationally designed two-fold symmetric tim-barrelPDBTitle: computationally designed tim-barrel protein, halfflr
47 d1dkua1
not modelled
73.6
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like48 d1ovma1
not modelled
72.6
16
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain49 c3fhwB_
not modelled
72.4
13
PDB header: dna binding proteinChain: B: PDB Molecule: primosomal replication protein n;PDBTitle: crystal structure of the protein prib from bordetella parapertussis.2 northeast structural genomics consortium target bpr162.
50 c3guxA_
not modelled
72.1
20
PDB header: hydrolaseChain: A: PDB Molecule: putative zn-dependent exopeptidase;PDBTitle: crystal structure of a putative zn-dependent exopeptidase (bvu_1317)2 from bacteroides vulgatus atcc 8482 at 1.80 a resolution
51 c3ke8A_
not modelled
72.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphatePDBTitle: crystal structure of isph:hmbpp-complex
52 d1a9xb2
not modelled
71.9
15
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)53 c1keeH_
not modelled
71.7
15
PDB header: ligaseChain: H: PDB Molecule: carbamoyl-phosphate synthetase small chain;PDBTitle: inactivation of the amidotransferase activity of carbamoyl phosphate2 synthetase by the antibiotic acivicin
54 d1pqua1
not modelled
70.9
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain55 d1iuka_
not modelled
70.8
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain56 c2decA_
not modelled
70.8
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 325aa long hypothetical protein;PDBTitle: crystal structure of the ph0510 protein from pyrococcus horikoshii ot3
57 d1thfd_
not modelled
70.2
15
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes58 c3k8aA_
not modelled
69.5
13
PDB header: dna binding proteinChain: A: PDB Molecule: putative primosomal replication protein;PDBTitle: neisseria gonorrhoeae prib
59 c2ivfA_
not modelled
68.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: ethylbenzene dehydrogenase alpha-subunit;PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
60 c3izbA_
not modelled
68.4
19
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein rps0 (s2p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
61 d1vlva2
not modelled
68.3
26
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase62 c3olhA_
not modelled
67.9
20
PDB header: transferaseChain: A: PDB Molecule: 3-mercaptopyruvate sulfurtransferase;PDBTitle: human 3-mercaptopyruvate sulfurtransferase
63 d2ji7a1
not modelled
67.4
10
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain64 c2v45A_
not modelled
67.2
11
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic nitrate reductase;PDBTitle: a new catalytic mechanism of periplasmic nitrate reductase2 from desulfovibrio desulfuricans atcc 27774 from3 crystallographic and epr data and based on detailed4 analysis of the sixth ligand
65 c2x3yA_
not modelled
66.6
14
PDB header: isomeraseChain: A: PDB Molecule: phosphoheptose isomerase;PDBTitle: crystal structure of gmha from burkholderia pseudomallei
66 c3eafA_
not modelled
66.3
11
PDB header: transport proteinChain: A: PDB Molecule: abc transporter, substrate binding protein;PDBTitle: crystal structure of abc transporter, substrate binding protein2 aeropyrum pernix
67 c3fxaA_
not modelled
66.0
15
PDB header: sugar binding proteinChain: A: PDB Molecule: sis domain protein;PDBTitle: crystal structure of a putative sugar-phosphate isomerase2 (lmof2365_0531) from listeria monocytogenes str. 4b f2365 at 1.60 a3 resolution
68 d2jioa2
not modelled
66.0
10
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-369 d1gm5a2
not modelled
65.8
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RecG "wedge" domain70 c3gdwA_
not modelled
65.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sigma-54 interaction domain protein;PDBTitle: crystal structure of sigma-54 interaction domain protein from2 enterococcus faecalis
71 c2xhzC_
not modelled
64.6
15
PDB header: isomeraseChain: C: PDB Molecule: arabinose 5-phosphate isomerase;PDBTitle: probing the active site of the sugar isomerase domain from e. coli2 arabinose-5-phosphate isomerase via x-ray crystallography
72 c3cwoX_
not modelled
64.3
21
PDB header: de novo proteinChain: X: PDB Molecule: beta/alpha-barrel protein based on 1thf and 1tmy;PDBTitle: a beta/alpha-barrel built by the combination of fragments2 from different folds
73 d1u9ya2
not modelled
64.1
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like74 d1jeoa_
not modelled
64.0
11
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain75 c1z9fA_
not modelled
63.6
7
PDB header: dna binding proteinChain: A: PDB Molecule: single-strand binding protein;PDBTitle: crystal structure of single stranded dna-binding protein (tm0604) from2 thermotoga maritima at 2.60 a resolution
76 d1urha2
not modelled
63.2
16
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)77 c3h75A_
not modelled
62.8
14
PDB header: sugar binding proteinChain: A: PDB Molecule: periplasmic sugar-binding domain protein;PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
78 c2y85D_
not modelled
62.8
17
PDB header: isomeraseChain: D: PDB Molecule: phosphoribosyl isomerase a;PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
79 d1ka9f_
not modelled
62.4
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes80 d1eyea_
not modelled
62.3
19
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase81 c1dkrB_
not modelled
62.1
11
PDB header: transferaseChain: B: PDB Molecule: phosphoribosyl pyrophosphate synthetase;PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
82 c3shoA_
not modelled
61.8
17
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, rpir family;PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
83 c3qy6A_
not modelled
61.8
37
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase ywqe;PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
84 d1ml4a2
not modelled
61.4
14
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase85 d1o6ca_
not modelled
61.4
15
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDP-N-acetylglucosamine 2-epimerase86 c1urhA_
not modelled
60.7
15
PDB header: transferaseChain: A: PDB Molecule: 3-mercaptopyruvate sulfurtransferase;PDBTitle: the "rhodanese" fold and catalytic mechanism of2 3-mercaptopyruvate sulfotransferases: crystal structure3 of ssea from escherichia coli
87 c2zkqb_
not modelled
60.1
18
PDB header: ribosomal protein/rnaChain: B: PDB Molecule: rna expansion segment es3;PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
88 c1s1hB_
not modelled
59.8
18
PDB header: ribosomeChain: B: PDB Molecule: 40s ribosomal protein s0-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
89 d1edga_
not modelled
59.7
22
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases90 c3lpnB_
not modelled
59.1
19
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase;PDBTitle: crystal structure of the phosphoribosylpyrophosphate (prpp) synthetase2 from thermoplasma volcanium in complex with an atp analog (ampcpp).
91 c3iz6A_
not modelled
59.0
17
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein sa (s2p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
92 d1ozha1
not modelled
58.7
9
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain93 d2ihta1
not modelled
58.5
16
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain94 c1yz4A_
not modelled
58.3
12
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase-like 15 isoform a;PDBTitle: crystal structure of dusp15
95 c1boiA_
not modelled
57.9
13
PDB header: transferaseChain: A: PDB Molecule: rhodanese;PDBTitle: n-terminally truncated rhodanese
96 c2yybA_
not modelled
57.9
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1606;PDBTitle: crystal structure of ttha1606 from thermus thermophilus hb8
97 c3nuhB_
not modelled
57.8
25
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit b;PDBTitle: a domain insertion in e. coli gyrb adopts a novel fold that plays a2 critical role in gyrase function
98 d2pi2a1
not modelled
57.2
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB99 d1ybha1
not modelled
57.2
9
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain100 c3outC_
not modelled
57.0
12
PDB header: isomeraseChain: C: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
101 d1ecfa1
not modelled
57.0
23
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)102 d1c4oa2
not modelled
56.9
8
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain103 d1pq4a_
not modelled
56.6
15
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like104 c2puwA_
not modelled
56.5
10
PDB header: transferaseChain: A: PDB Molecule: isomerase domain of glutamine-fructose-6-phosphatePDBTitle: the crystal structure of isomerase domain of glucosamine-6-phosphate2 synthase from candida albicans
105 c2xznB_
not modelled
56.4
17
PDB header: ribosomeChain: B: PDB Molecule: rps0e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
106 c3dmdA_
not modelled
55.8
17
PDB header: transport proteinChain: A: PDB Molecule: signal recognition particle receptor;PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
107 c2pi2A_
not modelled
55.7
18
PDB header: replication, dna binding proteinChain: A: PDB Molecule: replication protein a 32 kda subunit;PDBTitle: full-length replication protein a subunits rpa14 and rpa32
108 c3aaxB_
not modelled
55.6
10
PDB header: transferaseChain: B: PDB Molecule: putative thiosulfate sulfurtransferase;PDBTitle: crystal structure of probable thiosulfate sulfurtransferase2 cysa3 (rv3117) from mycobacterium tuberculosis: monoclinic3 form
109 d1otha2
not modelled
55.4
27
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase110 c1y5iA_
not modelled
55.3
13
PDB header: oxidoreductaseChain: A: PDB Molecule: respiratory nitrate reductase 1 alpha chain;PDBTitle: the crystal structure of the narghi mutant nari-k86a
111 c2ph5A_
not modelled
55.2
19
PDB header: transferaseChain: A: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
112 c3rlhA_
not modelled
55.1
23
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelin phosphodiesterase d lisictox-alphaia1a;PDBTitle: crystal structure of a class ii phospholipase d from loxosceles2 intermedia venom
113 c3eyxB_
not modelled
54.9
24
PDB header: lyaseChain: B: PDB Molecule: carbonic anhydrase;PDBTitle: crystal structure of carbonic anhydrase nce103 from2 saccharomyces cerevisiae
114 c2yz5B_
not modelled
54.0
21
PDB header: hydrolaseChain: B: PDB Molecule: histidinol phosphatase;PDBTitle: histidinol phosphate phosphatase complexed with phosphate
115 d1duvg2
not modelled
53.9
18
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase116 c3q41B_
not modelled
53.7
12
PDB header: transport proteinChain: B: PDB Molecule: glutamate [nmda] receptor subunit zeta-1;PDBTitle: crystal structure of the glun1 n-terminal domain (ntd)
117 d1m65a_
not modelled
53.3
28
Fold: 7-stranded beta/alpha barrelSuperfamily: PHP domain-likeFamily: PHP domain118 c2j17A_
not modelled
53.2
16
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase yil113w;PDBTitle: ptyr bound form of sdp-1
119 c2w3nA_
not modelled
53.1
29
PDB header: lyaseChain: A: PDB Molecule: carbonic anhydrase 2;PDBTitle: structure and inhibition of the co2-sensing carbonic2 anhydrase can2 from the pathogenic fungus cryptococcus3 neoformans
120 c3pshA_
not modelled
52.7
10
PDB header: metal transportChain: A: PDB Molecule: protein hi_1472;PDBTitle: classification of a haemophilus influenzae abc transporter hi1470/712 through its cognate molybdate periplasmic binding protein mola (mola3 bound to molybdate)