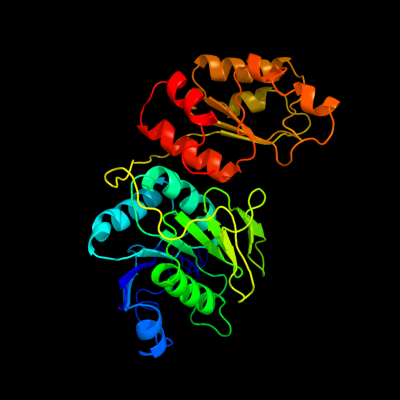
| 1 | d2ocda1
|
|
 |
100.0 |
69 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
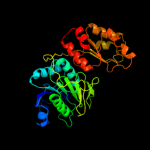
| 2 | c2d6fA_
|
|
 |
100.0 |
31 |
PDB header:ligase/rna
Chain: A: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: crystal structure of glu-trna(gln) amidotransferase in the2 complex with trna(gln)
|
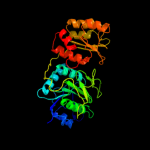
| 3 | c1zq1B_
|
|
 |
100.0 |
30 |
PDB header:lyase
Chain: B: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
|
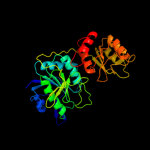
| 4 | d1zq1a2
|
|
 |
100.0 |
30 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 5 | c1wnfA_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of ph0066 from pyrococcus horikoshii
|
| 6 | d2d6fa2
|
|
 |
100.0 |
30 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 7 | d1nnsa_
|
|
 |
100.0 |
25 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 8 | d1wsaa_
|
|
 |
100.0 |
24 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 9 | c2wltA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: the crystal structure of helicobacter pylori l-asparaginase2 at 1.4 a resolution
|
| 10 | d1agxa_
|
|
 |
100.0 |
20 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 11 | d1o7ja_
|
|
 |
100.0 |
20 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 12 | d4pgaa_
|
|
 |
100.0 |
20 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 13 | c3nxkE_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: E: PDB Molecule:cytoplasmic l-asparaginase;
PDBTitle: crystal structure of probable cytoplasmic l-asparaginase from2 campylobacter jejuni
|
| 14 | c1uz5A_
|
|
 |
91.0 |
22 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
| 15 | d1gtza_
|
|
 |
90.2 |
18 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 16 | c2nqqA_
|
|
 |
86.8 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
| 17 | d1uz5a3
|
|
 |
86.3 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 18 | d1gqoa_
|
|
 |
83.4 |
14 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 19 | c2rjoA_
|
|
 |
81.2 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:twin-arginine translocation pathway signal protein;
PDBTitle: crystal structure of twin-arginine translocation pathway signal2 protein from burkholderia phytofirmans
|
| 20 | c2q62A_
|
|
 |
79.0 |
12 |
PDB header:flavoprotein
Chain: A: PDB Molecule:arsh;
PDBTitle: crystal structure of arsh from sinorhizobium meliloti
|
| 21 | d2ftsa3 |
|
not modelled |
76.6 |
11 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 22 | d2nqra3 |
|
not modelled |
74.7 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 23 | c2pr7A_ |
|
not modelled |
74.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase/epoxide hydrolase family;
PDBTitle: crystal structure of uncharacterized protein (np_599989.1) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.44 a resolution
|
| 24 | c3s81A_ |
|
not modelled |
73.6 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
|
| 25 | d1wu2a3 |
|
not modelled |
72.8 |
20 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 26 | c1powA_ |
|
not modelled |
72.2 |
13 |
PDB header:oxidoreductase(oxygen as acceptor)
Chain: A: PDB Molecule:pyruvate oxidase;
PDBTitle: the refined structures of a stabilized mutant and of wild-type2 pyruvate oxidase from lactobacillus plantarum
|
| 27 | d1goxa_ |
|
not modelled |
71.3 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 28 | c3d54D_ |
|
not modelled |
70.9 |
25 |
PDB header:ligase
Chain: D: PDB Molecule:phosphoribosylformylglycinamidine synthase 1;
PDBTitle: stucture of purlqs from thermotoga maritima
|
| 29 | d1tuba1 |
|
not modelled |
70.1 |
13 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 30 | c2fu3A_ |
|
not modelled |
68.1 |
9 |
PDB header:biosynthetic protein/structural protein
Chain: A: PDB Molecule:gephyrin;
PDBTitle: crystal structure of gephyrin e-domain
|
| 31 | c3ffsC_ |
|
not modelled |
67.3 |
5 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: the crystal structure of cryptosporidium parvum inosine-5'-2 monophosphate dehydrogenase
|
| 32 | c3o8nA_ |
|
not modelled |
67.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:6-phosphofructokinase, muscle type;
PDBTitle: structure of phosphofructokinase from rabbit skeletal muscle
|
| 33 | d2ioja1 |
|
not modelled |
66.7 |
32 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:DRTGG domain |
| 34 | d1h05a_ |
|
not modelled |
66.2 |
24 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 35 | d1tb3a1 |
|
not modelled |
65.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 36 | c1wu2B_ |
|
not modelled |
65.3 |
18 |
PDB header:structural genomics,biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis moea protein;
PDBTitle: crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikoshii ot3
|
| 37 | d1uqra_ |
|
not modelled |
64.2 |
18 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 38 | c3n8kG_ |
|
not modelled |
63.0 |
24 |
PDB header:lyase
Chain: G: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: type ii dehydroquinase from mycobacterium tuberculosis complexed with2 citrazinic acid
|
| 39 | c2cdh1_ |
|
not modelled |
62.0 |
20 |
PDB header:transferase
Chain: 1: PDB Molecule:enoyl reductase;
PDBTitle: architecture of the thermomyces lanuginosus fungal fatty2 acid synthase at 5 angstrom resolution.
|
| 40 | c2z6jB_ |
|
not modelled |
61.6 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trans-2-enoyl-acp reductase ii;
PDBTitle: crystal structure of s. pneumoniae enoyl-acyl carrier2 protein reductase (fabk) in complex with an inhibitor
|
| 41 | d1dxya2 |
|
not modelled |
61.5 |
18 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 42 | d1s1ma1 |
|
not modelled |
60.8 |
13 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 43 | c2hmcA_ |
|
not modelled |
60.0 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
| 44 | d1q7ra_ |
|
not modelled |
57.0 |
18 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 45 | c1jcnA_ |
|
not modelled |
56.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase i;
PDBTitle: binary complex of human type-i inosine monophosphate dehydrogenase2 with 6-cl-imp
|
| 46 | c3o1hB_ |
|
not modelled |
55.6 |
11 |
PDB header:signaling protein
Chain: B: PDB Molecule:periplasmic protein tort;
PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
|
| 47 | d1fuia2 |
|
not modelled |
55.6 |
9 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:L-fucose isomerase, N-terminal and second domains |
| 48 | d1ka9h_ |
|
not modelled |
55.0 |
18 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 49 | c2zskA_ |
|
not modelled |
54.8 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:226aa long hypothetical aspartate racemase;
PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
|
| 50 | c3brsA_ |
|
not modelled |
54.2 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of sugar transporter from clostridium2 phytofermentans
|
| 51 | c2dgdD_ |
|
not modelled |
53.6 |
12 |
PDB header:lyase
Chain: D: PDB Molecule:223aa long hypothetical arylmalonate decarboxylase;
PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
|
| 52 | c3mc3A_ |
|
not modelled |
53.0 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsre/dsrf-like family protein;
PDBTitle: crystal structure of dsre/dsrf-like family protein (np_342589.1) from2 sulfolobus solfataricus at 1.49 a resolution
|
| 53 | c2e77B_ |
|
not modelled |
52.4 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate oxidase;
PDBTitle: crystal structure of l-lactate oxidase with pyruvate complex
|
| 54 | d1ccwa_ |
|
not modelled |
52.2 |
17 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 55 | d1xi8a3 |
|
not modelled |
52.0 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 56 | d1jlja_ |
|
not modelled |
51.6 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 57 | d1i1qb_ |
|
not modelled |
51.4 |
14 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 58 | c3bo9B_ |
|
not modelled |
51.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nitroalkan dioxygenase;
PDBTitle: crystal structure of putative nitroalkan dioxygenase (tm0800) from2 thermotoga maritima at 2.71 a resolution
|
| 59 | c2rduA_ |
|
not modelled |
50.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyacid oxidase 1;
PDBTitle: crystal structure of human glycolate oxidase in complex with2 glyoxylate
|
| 60 | c3lloA_ |
|
not modelled |
50.5 |
18 |
PDB header:motor protein
Chain: A: PDB Molecule:prestin;
PDBTitle: crystal structure of the stas domain of motor protein prestin (anion2 transporter slc26a5)
|
| 61 | c1jscA_ |
|
not modelled |
50.4 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:acetohydroxy-acid synthase;
PDBTitle: crystal structure of the catalytic subunit of yeast2 acetohydroxyacid synthase: a target for herbicidal3 inhibitors
|
| 62 | d2nv0a1 |
|
not modelled |
50.1 |
25 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 63 | c3lwzC_ |
|
not modelled |
50.0 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-2 dehydroquinate dehydratase (aroq) from yersinia pestis
|
| 64 | d2csua3 |
|
not modelled |
49.8 |
15 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 65 | c2qs0A_ |
|
not modelled |
49.7 |
25 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:quinolinate synthetase a;
PDBTitle: quinolinate synthase from pyrococcus furiosus
|
| 66 | d1wl8a1 |
|
not modelled |
49.5 |
14 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 67 | c2xecD_ |
|
not modelled |
49.4 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 68 | d1vcoa1 |
|
not modelled |
49.4 |
20 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 69 | d2c4va1 |
|
not modelled |
49.0 |
33 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 70 | c3khjE_ |
|
not modelled |
48.9 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
|
| 71 | d2csua1 |
|
not modelled |
48.8 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 72 | c3nurA_ |
|
not modelled |
48.5 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidohydrolase;
PDBTitle: crystal structure of a putative amidohydrolase from staphylococcus2 aureus
|
| 73 | c2opiB_ |
|
not modelled |
48.5 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:l-fuculose-1-phosphate aldolase;
PDBTitle: crystal structure of l-fuculose-1-phosphate aldolase from bacteroides2 thetaiotaomicron
|
| 74 | d1t3ta2 |
|
not modelled |
48.0 |
18 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 75 | c2r94B_ |
|
not modelled |
48.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 76 | d2btoa1 |
|
not modelled |
47.9 |
15 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 77 | d2hbva1 |
|
not modelled |
47.1 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:PP1699/LP2961-like |
| 78 | c2iexA_ |
|
not modelled |
47.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxynapthoic acid synthetase;
PDBTitle: crystal structure of dihydroxynapthoic acid synthetase (gk2873) from2 geobacillus kaustophilus hta426
|
| 79 | d1j4aa2 |
|
not modelled |
46.7 |
18 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 80 | d1q8fa_ |
|
not modelled |
46.5 |
20 |
Fold:Nucleoside hydrolase
Superfamily:Nucleoside hydrolase
Family:Nucleoside hydrolase |
| 81 | d1r61a_ |
|
not modelled |
45.4 |
10 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Putative cyclase
Family:Putative cyclase |
| 82 | d1gpma2 |
|
not modelled |
44.4 |
16 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 83 | c3ojcD_ |
|
not modelled |
43.8 |
9 |
PDB header:isomerase
Chain: D: PDB Molecule:putative aspartate/glutamate racemase;
PDBTitle: crystal structure of a putative asp/glu racemase from yersinia pestis
|
| 84 | c2uygF_ |
|
not modelled |
43.2 |
19 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
|
| 85 | d1uuya_ |
|
not modelled |
43.1 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 86 | d1y5ea1 |
|
not modelled |
42.0 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 87 | c2obnA_ |
|
not modelled |
41.8 |
24 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a duf1611 family protein (ava_3511) from anabaena2 variabilis atcc 29413 at 2.30 a resolution
|
| 88 | c2fzvC_ |
|
not modelled |
41.5 |
11 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative arsenical resistance protein;
PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
|
| 89 | c2vlbC_ |
|
not modelled |
41.4 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 90 | c1ozhD_ |
|
not modelled |
41.0 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:acetolactate synthase, catabolic;
PDBTitle: the crystal structure of klebsiella pneumoniae acetolactate2 synthase with enzyme-bound cofactor and with an unusual3 intermediate.
|
| 91 | c2issF_ |
|
not modelled |
40.9 |
27 |
PDB header:lyase, transferase
Chain: F: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: structure of the plp synthase holoenzyme from thermotoga maritima
|
| 92 | d2dlda2 |
|
not modelled |
40.8 |
25 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 93 | c3l7nA_ |
|
not modelled |
39.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of smu.1228c
|
| 94 | c2a7rD_ |
|
not modelled |
39.3 |
10 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gmp reductase 2;
PDBTitle: crystal structure of human guanosine monophosphate2 reductase 2 (gmpr2)
|
| 95 | d1p4ca_ |
|
not modelled |
38.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 96 | d1wzua1 |
|
not modelled |
38.9 |
35 |
Fold:NadA-like
Superfamily:NadA-like
Family:NadA-like |
| 97 | d1q6za1 |
|
not modelled |
38.4 |
12 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 98 | c3kipU_ |
|
not modelled |
38.1 |
24 |
PDB header:lyase
Chain: U: PDB Molecule:3-dehydroquinase, type ii;
PDBTitle: crystal structure of type-ii 3-dehydroquinase from c. albicans
|
| 99 | c3g1wB_ |
|
not modelled |
37.8 |
13 |
PDB header:transport protein
Chain: B: PDB Molecule:sugar abc transporter;
PDBTitle: crystal structure of sugar abc transporter (sugar-binding protein)2 from bacillus halodurans
|
| 100 | c2a7nA_ |
|
not modelled |
37.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l(+)-mandelate dehydrogenase;
PDBTitle: crystal structure of the g81a mutant of the active chimera of (s)-2 mandelate dehydrogenase
|
| 101 | d1jcna1 |
|
not modelled |
37.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 102 | c1brwB_ |
|
not modelled |
37.2 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyrimidine nucleoside phosphorylase);
PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
|
| 103 | d1mx3a2 |
|
not modelled |
37.1 |
18 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 104 | c2htmB_ |
|
not modelled |
37.0 |
16 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiazole biosynthesis protein thig;
PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
|
| 105 | d1k9vf_ |
|
not modelled |
36.9 |
23 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 106 | d1vmea1 |
|
not modelled |
36.2 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 107 | d1qwga_ |
|
not modelled |
36.2 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
| 108 | d1eepa_ |
|
not modelled |
36.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 109 | c2qneA_ |
|
not modelled |
36.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase;
PDBTitle: crystal structure of putative methyltransferase (zp_00558420.1) from2 desulfitobacterium hafniense y51 at 2.30 a resolution
|
| 110 | d1ls1a2 |
|
not modelled |
36.0 |
22 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 111 | c1keeH_ |
|
not modelled |
35.5 |
17 |
PDB header:ligase
Chain: H: PDB Molecule:carbamoyl-phosphate synthetase small chain;
PDBTitle: inactivation of the amidotransferase activity of carbamoyl phosphate2 synthetase by the antibiotic acivicin
|
| 112 | c3gdwA_ |
|
not modelled |
35.5 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:sigma-54 interaction domain protein;
PDBTitle: crystal structure of sigma-54 interaction domain protein from2 enterococcus faecalis
|
| 113 | d1tubb1 |
|
not modelled |
35.2 |
17 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 114 | c3bezC_ |
|
not modelled |
35.2 |
28 |
PDB header:hydrolase
Chain: C: PDB Molecule:protease 4;
PDBTitle: crystal structure of escherichia coli signal peptide peptidase (sppa),2 semet crystals
|
| 115 | d1jr1a1 |
|
not modelled |
34.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 116 | c2ax3A_ |
|
not modelled |
34.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein tm0922;
PDBTitle: crystal structure of a putative carbohydrate kinase (tm0922) from2 thermotoga maritima msb8 at 2.25 a resolution
|
| 117 | d1im5a_ |
|
not modelled |
34.0 |
16 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 118 | c3k2qA_ |
|
not modelled |
33.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:pyrophosphate-dependent phosphofructokinase;
PDBTitle: crystal structure of pyrophosphate-dependent2 phosphofructokinase from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr88
|
| 119 | c3ij6A_ |
|
not modelled |
33.6 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized metal-dependent hydrolase;
PDBTitle: crystal structure of an uncharacterized metal-dependent2 hydrolase from lactobacillus acidophilus
|
| 120 | d1ezra_ |
|
not modelled |
33.5 |
12 |
Fold:Nucleoside hydrolase
Superfamily:Nucleoside hydrolase
Family:Nucleoside hydrolase |