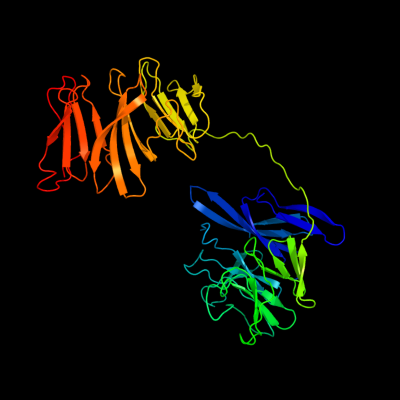
1 c1k32E_
100.0
10
PDB header: hydrolaseChain: E: PDB Molecule: tricorn protease;PDBTitle: crystal structure of the tricorn protease
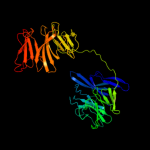
2 c1n6dE_
100.0
10
PDB header: hydrolaseChain: E: PDB Molecule: tricorn protease;PDBTitle: tricorn protease in complex with tetrapeptide chloromethyl2 ketone derivative
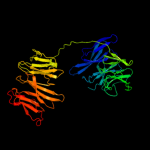
3 c1nr0A_
100.0
12
PDB header: structural proteinChain: A: PDB Molecule: actin interacting protein 1;PDBTitle: two seven-bladed beta-propeller domains revealed by the2 structure of a c. elegans homologue of yeast actin3 interacting protein 1 (aip1).
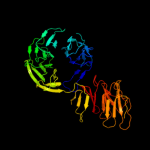
4 c3pe7A_
100.0
13
PDB header: lyaseChain: A: PDB Molecule: oligogalacturonate lyase;PDBTitle: oligogalacturonate lyase in complex with manganese
5 c3iytG_
100.0
16
PDB header: apoptosisChain: G: PDB Molecule: apoptotic protease-activating factor 1;PDBTitle: structure of an apoptosome-procaspase-9 card complex
6 d1k32a3
100.0
10
Fold: 7-bladed beta-propellerSuperfamily: Tricorn protease domain 2Family: Tricorn protease domain 27 c3dm0A_
100.0
13
PDB header: sugar binding protein,signaling proteinChain: A: PDB Molecule: maltose-binding periplasmic protein fused withPDBTitle: maltose binding protein fusion with rack1 from a. thaliana
8 c1pi6A_
99.9
14
PDB header: protein bindingChain: A: PDB Molecule: actin interacting protein 1;PDBTitle: yeast actin interacting protein 1 (aip1), orthorhombic crystal form
9 d1xfda1
99.9
11
Fold: 8-bladed beta-propellerSuperfamily: DPP6 N-terminal domain-likeFamily: DPP6 N-terminal domain-like10 c2w8bB_
99.9
13
PDB header: protein transport/membrane proteinChain: B: PDB Molecule: protein tolb;PDBTitle: crystal structure of processed tolb in complex with pal
11 c3c5mC_
99.9
11
PDB header: lyaseChain: C: PDB Molecule: oligogalacturonate lyase;PDBTitle: crystal structure of oligogalacturonate lyase (vpa0088)2 from vibrio parahaemolyticus. northeast structural3 genomics consortium target vpr199
12 d1orva1
99.9
10
Fold: 8-bladed beta-propellerSuperfamily: DPP6 N-terminal domain-likeFamily: DPP6 N-terminal domain-like13 c3bwsA_
99.9
11
PDB header: unknown functionChain: A: PDB Molecule: protein lp49;PDBTitle: crystal structure of the leptospiral antigen lp49
14 d2bgra1
99.9
9
Fold: 8-bladed beta-propellerSuperfamily: DPP6 N-terminal domain-likeFamily: DPP6 N-terminal domain-like15 d1nira2
99.9
10
Fold: 8-bladed beta-propellerSuperfamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductaseFamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductase16 c2ivzD_
99.9
12
PDB header: protein transport/hydrolaseChain: D: PDB Molecule: protein tolb;PDBTitle: structure of tolb in complex with a peptide of the colicin2 e9 t-domain
17 c1gq1B_
99.9
13
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome cd1 nitrite reductase;PDBTitle: cytochrome cd1 nitrite reductase, y25s mutant, oxidised2 form
18 c2oajA_
99.9
14
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: protein sni1;PDBTitle: crystal structure of sro7 from s. cerevisiae
19 c3jroA_
99.9
13
PDB header: transport protein, structural proteinChain: A: PDB Molecule: fusion protein of protein transport protein sec13PDBTitle: nup84-nup145c-sec13 edge element of the npc lattice
20 d1qksa2
99.9
10
Fold: 8-bladed beta-propellerSuperfamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductaseFamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductase21 c3mkqA_
not modelled
99.9
9
PDB header: transport proteinChain: A: PDB Molecule: coatomer beta'-subunit;PDBTitle: crystal structure of yeast alpha/betaprime-cop subcomplex of the copi2 vesicular coat
22 c1nnoA_
not modelled
99.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: nitrite reductase;PDBTitle: conformational changes occurring upon no binding in nitrite2 reductase from pseudomonas aeruginosa
23 d1gxra_
not modelled
99.9
13
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat24 d1jmxb_
not modelled
99.9
13
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Quinohemoprotein amine dehydrogenase B chain25 c1xfdD_
not modelled
99.9
11
PDB header: membrane proteinChain: D: PDB Molecule: dipeptidyl aminopeptidase-like protein 6;PDBTitle: structure of a human a-type potassium channel accelerating factor2 dppx, a member of the dipeptidyl aminopeptidase family
26 c2ecfA_
not modelled
99.9
15
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl peptidase iv;PDBTitle: crystal structure of dipeptidyl aminopeptidase iv from2 stenotrophomonas maltophilia
27 c1z68A_
not modelled
99.9
12
PDB header: lyaseChain: A: PDB Molecule: fibroblast activation protein, alpha subunit;PDBTitle: crystal structure of human fibroblast activation protein alpha
28 c2pm9A_
not modelled
99.9
13
PDB header: protein transportChain: A: PDB Molecule: protein transport protein sec31;PDBTitle: crystal structure of yeast sec13/31 vertex element of the2 copii vesicular coat
29 c2qtbB_
not modelled
99.9
9
PDB header: hydrolaseChain: B: PDB Molecule: dipeptidyl peptidase 4;PDBTitle: human dipeptidyl peptidase iv/cd26 in complex with a 4-aryl2 cyclohexylalanine inhibitor
30 c2g5tA_
not modelled
99.9
8
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl peptidase 4;PDBTitle: crystal structure of human dipeptidyl peptidase iv (dppiv)2 complexed with cyanopyrrolidine (c5-pro-pro) inhibitor 21ag
31 c3fgbB_
not modelled
99.9
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein q89zh8_bactn;PDBTitle: crystal structure of the q89zh8_bactn protein from2 bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr289b.
32 c1r5mA_
not modelled
99.9
7
PDB header: transcriptionChain: A: PDB Molecule: sir4-interacting protein sif2;PDBTitle: crystal structure of the c-terminal wd40 domain of sif2
33 c2eepA_
not modelled
99.9
14
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl aminopeptidase iv, putative;PDBTitle: prolyl tripeptidyl aminopeptidase complexed with an inhibitor
34 c3ei4D_
not modelled
99.9
15
PDB header: dna binding proteinChain: D: PDB Molecule: dna damage-binding protein 2;PDBTitle: structure of the hsddb1-hsddb2 complex
35 d1k32a2
not modelled
99.9
14
Fold: 6-bladed beta-propellerSuperfamily: Tricorn protease N-terminal domainFamily: Tricorn protease N-terminal domain36 c2j04B_
not modelled
99.9
12
PDB header: transcriptionChain: B: PDB Molecule: ydr362cp;PDBTitle: the tau60-tau91 subcomplex of yeast transcription factor2 iiic
37 c2j57J_
not modelled
99.9
13
PDB header: oxidoreductaseChain: J: PDB Molecule: methylamine dehydrogenase heavy chain;PDBTitle: x-ray reduced paraccocus denitrificans methylamine2 dehydrogenase n-quinol in complex with amicyanin.
38 c2i0tB_
not modelled
99.9
12
PDB header: oxidoreductaseChain: B: PDB Molecule: aromatic amine dehydrogenase;PDBTitle: crystal structure of phenylacetaldehyde derived r-2 carbinolamine adduct of aromatic amine dehydrogenase
39 d2hqsa1
not modelled
99.9
9
Fold: 6-bladed beta-propellerSuperfamily: TolB, C-terminal domainFamily: TolB, C-terminal domain40 c2h47F_
not modelled
99.9
13
PDB header: oxidoreductase/electron transportChain: F: PDB Molecule: aromatic amine dehydrogenase;PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
41 c3dw8B_
not modelled
99.8
11
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: serine/threonine-protein phosphatase 2a 55 kda regulatoryPDBTitle: structure of a protein phosphatase 2a holoenzyme with b55 subunit
42 c2gopB_
not modelled
99.8
9
PDB header: hydrolaseChain: B: PDB Molecule: trilobed protease;PDBTitle: the beta-propeller domain of the trilobed protease from pyrococcus2 furiosus reveals an open velcro topology
43 d1fwxa2
not modelled
99.8
7
Fold: 7-bladed beta-propellerSuperfamily: Nitrous oxide reductase, N-terminal domainFamily: Nitrous oxide reductase, N-terminal domain44 d1erja_
not modelled
99.8
12
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat45 c3ei3B_
not modelled
99.8
12
PDB header: dna binding proteinChain: B: PDB Molecule: dna damage-binding protein 2;PDBTitle: structure of the hsddb1-drddb2 complex
46 d1yfqa_
not modelled
99.8
12
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: Cell cycle arrest protein BUB347 c2ojhA_
not modelled
99.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1656/agr_c_3050;PDBTitle: the structure of putative tolb from agrobacterium tumefaciens
48 c2pbiB_
not modelled
99.8
14
PDB header: signaling proteinChain: B: PDB Molecule: guanine nucleotide-binding protein subunit beta 5;PDBTitle: the multifunctional nature of gbeta5/rgs9 revealed from its crystal2 structure
49 d2madh_
not modelled
99.8
12
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Methylamine dehydrogenase, H-chain50 d2bbkh_
not modelled
99.8
12
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Methylamine dehydrogenase, H-chain51 c3c75J_
not modelled
99.8
10
PDB header: oxidoreductaseChain: J: PDB Molecule: methylamine dehydrogenase heavy chain;PDBTitle: paracoccus versutus methylamine dehydrogenase in complex2 with amicyanin
52 c3acpA_
not modelled
99.8
11
PDB header: chaperoneChain: A: PDB Molecule: wd repeat-containing protein ygl004c;PDBTitle: crystal structure of yeast rpn14, a chaperone of the 19s regulatory2 particle of the proteasome
53 c2w18A_
not modelled
99.8
11
PDB header: nuclear proteinChain: A: PDB Molecule: partner and localizer of brca2;PDBTitle: crystal structure of the c-terminal wd40 domain of human2 palb2
54 c1qniE_
not modelled
99.8
12
PDB header: oxidoreductaseChain: E: PDB Molecule: nitrous-oxide reductase;PDBTitle: crystal structure of nitrous oxide reductase from2 pseudomonas nautica, at 2.4a resolution
55 c3fm0A_
not modelled
99.8
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: protein ciao1;PDBTitle: crystal structure of wd40 protein ciao1
56 d1nr0a1
not modelled
99.8
15
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat57 c3i2nA_
not modelled
99.8
14
PDB header: transcriptionChain: A: PDB Molecule: wd repeat-containing protein 92;PDBTitle: crystal structure of wd40 repeats protein wdr92
58 d1ospo_
not modelled
99.8
6
Fold: open-sided beta-meanderSuperfamily: Outer surface proteinFamily: Outer surface protein59 d1tbga_
not modelled
99.8
11
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat60 d1qnia2
not modelled
99.8
11
Fold: 7-bladed beta-propellerSuperfamily: Nitrous oxide reductase, N-terminal domainFamily: Nitrous oxide reductase, N-terminal domain61 c3jzhA_
not modelled
99.8
11
PDB header: gene regulationChain: A: PDB Molecule: polycomb protein eed;PDBTitle: eed-h3k79me3
62 d1l0qa2
not modelled
99.8
13
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: YVTN repeat63 c2xznR_
not modelled
99.8
12
PDB header: ribosomeChain: R: PDB Molecule: rack1;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
64 d1nexb2
not modelled
99.8
12
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat65 c3u4yA_
not modelled
99.8
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of a functionally unknown protein (dtox_1751)2 from desulfotomaculum acetoxidans dsm 771.
66 c3hfqB_
not modelled
99.8
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein lp_2219;PDBTitle: crystal structure of the lp_2219 protein from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr118.
67 c2gnqA_
not modelled
99.8
13
PDB header: transcriptionChain: A: PDB Molecule: wd-repeat protein 5;PDBTitle: structure of wdr5
68 c3mmyE_
not modelled
99.8
9
PDB header: nuclear proteinChain: E: PDB Molecule: mrna export factor;PDBTitle: structural and functional analysis of the interaction between the2 nucleoporin nup98 and the mrna export factor rae1
69 c2qxvA_
not modelled
99.8
12
PDB header: gene regulationChain: A: PDB Molecule: embryonic ectoderm development;PDBTitle: structural basis of ezh2 recognition by eed
70 c3iz6a_
not modelled
99.8
11
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein sa (s2p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
71 c4a11B_
not modelled
99.8
12
PDB header: dna binding proteinChain: B: PDB Molecule: dna excision repair protein ercc-8;PDBTitle: structure of the hsddb1-hscsa complex
72 c3frxB_
not modelled
99.8
13
PDB header: signaling proteinChain: B: PDB Molecule: guanine nucleotide-binding protein subunit beta-PDBTitle: crystal structure of the yeast orthologue of rack1, asc1.
73 c3jrpA_
not modelled
99.8
7
PDB header: transport protein, structural proteinChain: A: PDB Molecule: fusion protein of protein transport protein sec13PDBTitle: sec13 with nup145c (aa109-179) insertion blade
74 d1pgua1
not modelled
99.8
12
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat75 d2ovrb2
not modelled
99.8
8
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat76 d1vyhc1
not modelled
99.7
7
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat77 c3eg6A_
not modelled
99.7
12
PDB header: protein bindingChain: A: PDB Molecule: wd repeat-containing protein 5;PDBTitle: structure of wdr5 bound to mll1 peptide
78 c1fwxB_
not modelled
99.7
10
PDB header: oxidoreductaseChain: B: PDB Molecule: nitrous oxide reductase;PDBTitle: crystal structure of nitrous oxide reductase from p. denitrificans
79 c1vyhT_
not modelled
99.7
7
PDB header: hydrolaseChain: T: PDB Molecule: platelet-activating factor acetylhydrolase ibPDBTitle: paf-ah holoenzyme: lis1/alfa2
80 c1nexD_
not modelled
99.7
11
PDB header: ligase, cell cycleChain: D: PDB Molecule: cdc4 protein;PDBTitle: crystal structure of scskp1-sccdc4-cpd peptide complex
81 d1k8kc_
not modelled
99.7
12
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat82 d1sq9a_
not modelled
99.7
10
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat83 c2ovqB_
not modelled
99.7
12
PDB header: transcription/cell cycleChain: B: PDB Molecule: f-box/wd repeat protein 7;PDBTitle: structure of the skp1-fbw7-cyclinedegc complex
84 c3dwlH_
not modelled
99.7
12
PDB header: structural proteinChain: H: PDB Molecule: actin-related protein 2/3 complex subunit 1;PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
85 d1pbyb_
not modelled
99.7
12
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Quinohemoprotein amine dehydrogenase B chain86 c2vduB_
not modelled
99.7
13
PDB header: transferaseChain: B: PDB Molecule: trna (guanine-n(7)-)-methyltransferase-PDBTitle: structure of trm8-trm82, the yeast trna m7g methylation2 complex
87 d1ri6a_
not modelled
99.7
12
Fold: 7-bladed beta-propellerSuperfamily: Putative isomerase YbhEFamily: Putative isomerase YbhE88 c3fhcA_
not modelled
99.7
12
PDB header: transport protein/hydrolaseChain: A: PDB Molecule: nuclear pore complex protein nup214;PDBTitle: crystal structure of human dbp5 in complex with nup214
89 c3vh0C_
not modelled
99.7
13
PDB header: protein binding/dnaChain: C: PDB Molecule: uncharacterized protein ynce;PDBTitle: crystal structure of e. coli ynce complexed with dna
90 d1tl2a_
not modelled
99.7
12
Fold: 5-bladed beta-propellerSuperfamily: Tachylectin-2Family: Tachylectin-291 c3ow8A_
not modelled
99.7
9
PDB header: transcriptionChain: A: PDB Molecule: wd repeat-containing protein 61;PDBTitle: crystal structure of the wd repeat-containing protein 61
92 d1qfma1
not modelled
99.7
12
Fold: 7-bladed beta-propellerSuperfamily: Peptidase/esterase 'gauge' domainFamily: Prolyl oligopeptidase, N-terminal domain93 c3g4hB_
not modelled
99.7
16
PDB header: hydrolaseChain: B: PDB Molecule: regucalcin;PDBTitle: crystal structure of human senescence marker protein-30 (zinc bound)
94 d1nr0a2
not modelled
99.7
8
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat95 c3greA_
not modelled
99.7
13
PDB header: signaling protein,protein bindingChain: A: PDB Molecule: serine/threonine-protein kinase vps15;PDBTitle: crystal structure of saccharomyces cerevisiae vps15 wd2 repeat domain
96 c3lrvA_
not modelled
99.7
11
PDB header: splicingChain: A: PDB Molecule: pre-mrna-splicing factor 19;PDBTitle: the prp19 wd40 domain contains a conserved protein interaction region2 essential for its function.
97 c2hu7A_
not modelled
99.6
20
PDB header: hydrolaseChain: A: PDB Molecule: acylamino-acid-releasing enzyme;PDBTitle: binding of inhibitors by acylaminoacyl peptidase
98 c1yr2A_
not modelled
99.6
13
PDB header: hydrolaseChain: A: PDB Molecule: prolyl oligopeptidase;PDBTitle: structural and mechanistic analysis of two prolyl endopeptidases: role2 of inter-domain dynamics in catalysis and specificity
99 c2aq5A_
not modelled
99.6
10
PDB header: structural proteinChain: A: PDB Molecule: coronin-1a;PDBTitle: crystal structure of murine coronin-1
100 c3odtB_
not modelled
99.6
7
PDB header: nuclear proteinChain: B: PDB Molecule: protein doa1;PDBTitle: crystal structure of wd40 beta propeller domain of doa1
101 c2bklB_
not modelled
99.6
13
PDB header: hydrolaseChain: B: PDB Molecule: prolyl endopeptidase;PDBTitle: structural and mechanistic analysis of two prolyl2 endopeptidases: role of inter-domain dynamics in3 catalysis and specificity
102 c3sbrF_
not modelled
99.6
11
PDB header: oxidoreductaseChain: F: PDB Molecule: nitrous-oxide reductase;PDBTitle: pseudomonas stutzeri nitrous oxide reductase, p1 crystal form with2 substrate
103 c1qfmA_
not modelled
99.5
13
PDB header: hydrolaseChain: A: PDB Molecule: protein (prolyl oligopeptidase);PDBTitle: prolyl oligopeptidase from porcine muscle
104 c3e5zA_
not modelled
99.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative gluconolactonase;PDBTitle: x-ray structure of the putative gluconolactonase in protein family2 pf08450. northeast structural genomics consortium target drr130.
105 c2g8sB_
not modelled
99.5
11
PDB header: sugar binding proteinChain: B: PDB Molecule: glucose/sorbosone dehydrogenases;PDBTitle: crystal structure of the soluble aldose sugar dehydrogenase2 (asd) from escherichia coli in the apo-form
106 c2hesX_
not modelled
99.5
11
PDB header: biosynthetic proteinChain: X: PDB Molecule: ydr267cp;PDBTitle: cytosolic iron-sulphur assembly protein- 1
107 c3cfvA_
not modelled
99.5
14
PDB header: histone/chaperoneChain: A: PDB Molecule: histone-binding protein rbbp7;PDBTitle: structural basis of the interaction of rbap46/rbap48 with2 histone h4
108 c2pm7B_
not modelled
99.5
14
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec13;PDBTitle: crystal structure of yeast sec13/31 edge element of the2 copii vesicular coat, selenomethionine version
109 c2zkqa_
not modelled
99.5
12
PDB header: ribosomal protein/rnaChain: A: PDB Molecule: 18s ribosomal rna;PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
110 c3bg1E_
not modelled
99.5
10
PDB header: protein transport, hydrolaseChain: E: PDB Molecule: protein sec13 homolog;PDBTitle: architecture of a coat for the nuclear pore membrane
111 c1p22A_
not modelled
99.4
9
PDB header: signaling proteinChain: A: PDB Molecule: f-box/wd-repeat protein 1a;PDBTitle: structure of a beta-trcp1-skp1-beta-catenin complex:2 destruction motif binding and lysine specificity on the3 scfbeta-trcp1 ubiquitin ligase
112 c2oitA_
not modelled
99.4
15
PDB header: oncoproteinChain: A: PDB Molecule: nucleoporin 214kda;PDBTitle: crystal structure of the n-terminal domain of the human2 proto-oncogene nup214/can
113 d2dg1a1
not modelled
99.4
9
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like114 c3azqA_
not modelled
99.4
15
PDB header: hydrolaseChain: A: PDB Molecule: aminopeptidase;PDBTitle: crystal structure of puromycin hydrolase s511a mutant complexed with2 pgg
115 c3eweC_
not modelled
99.4
9
PDB header: protein transport,structural proteinChain: C: PDB Molecule: nucleoporin seh1;PDBTitle: crystal structure of the nup85/seh1 complex
116 c3iumA_
not modelled
99.4
10
PDB header: hydrolaseChain: A: PDB Molecule: prolyl endopeptidase;PDBTitle: appep_wtx opened state
117 c3dr2A_
not modelled
99.4
13
PDB header: hydrolaseChain: A: PDB Molecule: exported gluconolactonase;PDBTitle: structural and functional analyses of xc5397 from2 xanthomonas campestris: a gluconolactonase important in3 glucose secondary metabolic pathways
118 d1pgua2
not modelled
99.3
10
Fold: 7-bladed beta-propellerSuperfamily: WD40 repeat-likeFamily: WD40-repeat119 d2p4oa1
not modelled
99.3
10
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: All0351-like120 d1pjxa_
not modelled
99.3
11
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like