| 1 | c3oyrB_
|
|
 |
100.0 |
45 |
PDB header:transferase
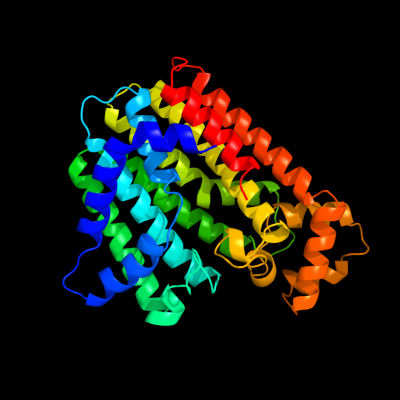
Chain: B: PDB Molecule:trans-isoprenyl diphosphate synthase;
PDBTitle: crystal structure of polyprenyl synthase from caulobacter crescentus2 cb15 complexed with calcium and isoprenyl diphosphate
|
| 2 | c3mzvB_
|
|
 |
100.0 |
46 |
PDB header:transferase
Chain: B: PDB Molecule:decaprenyl diphosphate synthase;
PDBTitle: crystal structure of a decaprenyl diphosphate synthase from2 rhodobacter capsulatus
|
| 3 | c1wy0A_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranyl pyrophosphate synthetase;
PDBTitle: crystal structure of geranylgeranyl pyrophosphate synthetase from2 pyrococcus horikoshii ot3
|
| 4 | c3aqbD_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: D: PDB Molecule:component b of hexaprenyl diphosphate synthase;
PDBTitle: m. luteus b-p 26 heterodimeric hexaprenyl diphosphate synthase in2 complex with magnesium
|
| 5 | c3nf2A_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:putative polyprenyl synthetase;
PDBTitle: crystal structure of polyprenyl synthetase from streptomyces2 coelicolor a3(2)
|
| 6 | c1wmwA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranyl diphosphate synthetase;
PDBTitle: crystal structure of geranulgeranyl diphosphate synthase from thermus2 thermophilus
|
| 7 | d1fpsa_
|
|
 |
100.0 |
22 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Isoprenyl diphosphate synthases |
| 8 | c3n3dB_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:geranylgeranyl pyrophosphate synthase;
PDBTitle: crystal structure of geranylgeranyl pyrophosphate synthase from2 lactobacillus brevis atcc 367
|
| 9 | c3aq0G_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: G: PDB Molecule:geranyl diphosphate synthase;
PDBTitle: ligand-bound form of arabidopsis medium/long-chain length prenyl2 pyrophosphate synthase (surface polar residue mutant)
|
| 10 | c3cp6A_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:farnesyl pyrophosphate synthetase;
PDBTitle: crystal structure of human farnesyl diphosphate synthase (t201a2 mutant) complexed with mg and biphosphonate inhibitor
|
| 11 | c3lmdA_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranyl pyrophosphate synthase;
PDBTitle: crystal structure of geranylgeranyl pyrophosphate synthase2 from corynebacterium glutamicum atcc 13032
|
| 12 | c2f94F_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: F: PDB Molecule:farnesyl diphosphate synthase;
PDBTitle: crystal structure of human fpps in complex with ibandronate
|
| 13 | d2q80a1
|
|
 |
100.0 |
22 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Isoprenyl diphosphate synthases |
| 14 | c3oyrA_
|
|
 |
100.0 |
45 |
PDB header:transferase
Chain: A: PDB Molecule:trans-isoprenyl diphosphate synthase;
PDBTitle: crystal structure of polyprenyl synthase from caulobacter crescentus2 cb15 complexed with calcium and isoprenyl diphosphate
|
| 15 | c2ogdB_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:farnesyl pyrophosphate synthase;
PDBTitle: t. brucei farnesyl diphosphate synthase complexed with bisphosphonate2 bph-527
|
| 16 | c3rmgB_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: B: PDB Molecule:octaprenyl-diphosphate synthase;
PDBTitle: crystal structure of geranylgeranyl pyrophosphate synthase from2 bacteroides thetaiotaomicron
|
| 17 | c1yhlA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:farnesyl pyrophosphate synthase;
PDBTitle: structure of the complex of trypanosoma cruzi farnesyl diphosphate2 synthase with risedronate, dmapp and mg+2
|
| 18 | c3ez3A_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:farnesyl pyrophosphate synthase, putative;
PDBTitle: crystal structure of plasmodium vivax geranylgeranylpyrophosphate2 synthase pvx_092040 with zoledronate and ipp bound
|
| 19 | d1rqja_
|
|
 |
100.0 |
31 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Isoprenyl diphosphate synthases |
| 20 | c2e8xB_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:geranylgeranyl pyrophosphate synthetase;
PDBTitle: s. cerevisiae geranylgeranyl pyrophosphate synthase in2 complex with magnesium and gpp
|
| 21 | d1v4ea_ |
|
not modelled |
100.0 |
27 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Isoprenyl diphosphate synthases |
| 22 | c3lk5A_ |
|
not modelled |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranyl pyrophosphate synthase;
PDBTitle: crystal structure of putative geranylgeranyl pyrophosphate synthase2 from corynebacterium glutamicum
|
| 23 | c2o1oB_ |
|
not modelled |
100.0 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative farnesyl pyrophosphate synthase;
PDBTitle: cryptosporidium parvum putative polyprenyl pyrophosphate2 synthase (cgd4_2550) in complex with risedronate.
|
| 24 | c3m9uD_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase
Chain: D: PDB Molecule:farnesyl-diphosphate synthase;
PDBTitle: crystal structure of geranylgeranyl pyrophosphate synthase2 from lactobacillus brevis atcc 367
|
| 25 | c3tc1A_ |
|
not modelled |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:octaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of octaprenyl pyrophosphate synthase from2 helicobacter pylori
|
| 26 | c3ts7B_ |
|
not modelled |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of farnesyl diphosphate synthase (target efi-501951)2 from methylococcus capsulatus
|
| 27 | d1rtra_ |
|
not modelled |
100.0 |
31 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Isoprenyl diphosphate synthases |
| 28 | c3ipiA_ |
|
not modelled |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of a geranyltranstransferase from the2 methanosarcina mazei
|
| 29 | c2forB_ |
|
not modelled |
100.0 |
31 |
PDB header:transferase
Chain: B: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of the shigella flexneri farnesyl pyrophosphate2 synthase complex with an isopentenyl pyrophosphate
|
| 30 | c2j1pB_ |
|
not modelled |
100.0 |
34 |
PDB header:transferase
Chain: B: PDB Molecule:geranylgeranyl pyrophosphate synthetase;
PDBTitle: geranylgeranyl diphosphate synthase from sinapis alba in2 complex with ggpp
|
| 31 | c3lsnA_ |
|
not modelled |
100.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of putative geranyltranstransferase from pseudomonas2 fluorescens pf-5 complexed with magnesium
|
| 32 | c3ucaB_ |
|
not modelled |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of isoprenoid synthase (target efi-501974) from2 clostridium perfringens
|
| 33 | c2h8oA_ |
|
not modelled |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase;
PDBTitle: the 1.6a crystal structure of the geranyltransferase from2 agrobacterium tumefaciens
|
| 34 | c2azjB_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:geranylgeranyl pyrophosphate synthetase;
PDBTitle: crystal structure for the mutant d81c of sulfolobus2 solfataricus hexaprenyl pyrophosphate synthase
|
| 35 | c3lomA_ |
|
not modelled |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of geranyltransferase from legionella2 pneumophila
|
| 36 | c3p8lB_ |
|
not modelled |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of polyprenyl synthase from enterococcus faecalis2 v583
|
| 37 | c3llwA_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase (ispa);
PDBTitle: crystal structure of geranyltransferase from helicobacter2 pylori 26695
|
| 38 | c3m0gB_ |
|
not modelled |
100.0 |
31 |
PDB header:transferase
Chain: B: PDB Molecule:farnesyl diphosphate synthase;
PDBTitle: crystal structure of putative farnesyl diphosphate synthase from2 rhodobacter capsulatus
|
| 39 | c2ftzA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of geranyltranstransferase (ec 2.5.1.10) (tm0161)2 from thermotoga maritima at 1.90 a resolution
|
| 40 | c2j1oA_ |
|
not modelled |
100.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranyl pyrophosphate synthetase;
PDBTitle: geranylgeranyl diphosphate synthase from sinapis alba
|
| 41 | c3qkcB_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:geranyl diphosphate synthase small subunit;
PDBTitle: crystal structure of geranyl diphosphate synthase small subunit from2 antirrhinum majus
|
| 42 | c3npkB_ |
|
not modelled |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:geranyltranstransferase;
PDBTitle: the crystal structure of geranyltranstransferase from campylobacter2 jejuni
|
| 43 | c3kraB_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:geranyl diphosphate synthase small subunit;
PDBTitle: mint heterotetrameric geranyl pyrophosphate synthase in2 complex with magnesium
|
| 44 | c3p8rA_ |
|
not modelled |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of polyprenyl synthase from vibrio cholerae
|
| 45 | d1ezfa_ |
|
not modelled |
98.0 |
13 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Squalene synthase |
| 46 | c2zcpA_ |
|
not modelled |
97.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dehydrosqualene synthase;
PDBTitle: crystal structure of the c(30) carotenoid dehydrosqualene2 synthase from staphylococcus aureus complexed with3 farnesyl thiopyrophosphate
|
| 47 | c3lg5A_ |
|
not modelled |
91.8 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:epi-isozizaene synthase;
PDBTitle: f198a epi-isozizaene synthase: complex with mg, inorganic2 pyrophosphate and benzyl triethyl ammonium cation
|
| 48 | d1ps1a_ |
|
not modelled |
86.5 |
13 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Aristolochene/pentalenene synthase |
| 49 | d1di1a_ |
|
not modelled |
86.4 |
13 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Aristolochene/pentalenene synthase |
| 50 | d1n1ba2 |
|
not modelled |
74.3 |
15 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Terpenoid cyclase C-terminal domain |
| 51 | c2oa6B_ |
|
not modelled |
61.7 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:aristolochene synthase;
PDBTitle: aristolochene synthase from aspergillus terreus complexed with2 pyrophosphate
|
| 52 | c1hm4A_ |
|
not modelled |
55.3 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:pentalenene synthase;
PDBTitle: n219l pentalenene synthase
|
| 53 | d2o3la1 |
|
not modelled |
41.9 |
25 |
Fold:Left-handed superhelix
Superfamily:BH3980-like
Family:BH3980-like |
| 54 | d2hh6a1 |
|
not modelled |
34.7 |
29 |
Fold:Left-handed superhelix
Superfamily:BH3980-like
Family:BH3980-like |
| 55 | c1hx9A_ |
|
not modelled |
34.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:5-epi-aristolochene synthase;
PDBTitle: crystal structure of teas w273s form 1
|
| 56 | c3n0fA_ |
|
not modelled |
26.5 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:isoprene synthase;
PDBTitle: crystal structure of isoprene synthase from grey poplar leaves2 (populus x canescens)
|
| 57 | c3g4dB_ |
|
not modelled |
24.5 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:(+)-delta-cadinene synthase isozyme xc1;
PDBTitle: crystal structure of (+)-delta-cadinene synthase from gossypium2 arboreum and evolutionary divergence of metal binding motifs for3 catalysis
|
| 58 | d1s7za_ |
|
not modelled |
20.1 |
21 |
Fold:Another 3-helical bundle
Superfamily:B-form DNA mimic Ocr
Family:B-form DNA mimic Ocr |
| 59 | c2ongA_ |
|
not modelled |
18.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:4s-limonene synthase;
PDBTitle: crystal structure of of limonene synthase with 2-2 fluorogeranyl diphosphate (fgpp).
|
| 60 | d5easa2 |
|
not modelled |
17.0 |
16 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Terpenoid cyclase C-terminal domain |
| 61 | c3byvA_ |
|
not modelled |
15.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:rhoptry kinase;
PDBTitle: crystal structure of toxoplasma gondii specific rhoptry2 antigen kinase domain
|
| 62 | c3p5rB_ |
|
not modelled |
15.4 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:taxadiene synthase;
PDBTitle: crystal structure of taxadiene synthase from pacific yew (taxus2 brevifolia) in complex with mg2+ and 2-fluorogeranylgeranyl3 diphosphate
|
| 63 | c3q60A_ |
|
not modelled |
13.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:rop5b;
PDBTitle: crystal structure of virulent allele rop5b pseudokinase domain bound2 to atp
|
| 64 | d2htja1 |
|
not modelled |
13.1 |
37 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:FaeA-like |
| 65 | d1ej5a_ |
|
not modelled |
11.5 |
15 |
Fold:Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Superfamily:Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Family:Wiscott-Aldrich syndrome protein, WASP, C-terminal domain |
| 66 | d1kiya_ |
|
not modelled |
10.7 |
9 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Trichodiene synthase |
| 67 | d2v4jc1 |
|
not modelled |
10.4 |
13 |
Fold:DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily:DsrC, the gamma subunit of dissimilatory sulfite reductase
Family:DsrC, the gamma subunit of dissimilatory sulfite reductase |
| 68 | c1n20A_ |
|
not modelled |
9.6 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:(+)-bornyl diphosphate synthase;
PDBTitle: (+)-bornyl diphosphate synthase: complex with mg and 3-aza-2 2,3-dihydrogeranyl diphosphate
|
| 69 | c3e7eA_ |
|
not modelled |
9.2 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:mitotic checkpoint serine/threonine-protein kinase bub1;
PDBTitle: structure and substrate recruitment of the human spindle checkpoint2 kinase bub
|
| 70 | c2jd3B_ |
|
not modelled |
8.8 |
19 |
PDB header:dna binding protein
Chain: B: PDB Molecule:stbb protein;
PDBTitle: parr from plasmid pb171
|
| 71 | c2v62A_ |
|
not modelled |
7.8 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine-protein kinase vrk2;
PDBTitle: structure of vaccinia-related kinase 2
|
| 72 | c3s9vD_ |
|
not modelled |
7.2 |
14 |
PDB header:lyase, isomerase
Chain: D: PDB Molecule:abietadiene synthase, chloroplastic;
PDBTitle: abietadiene synthase from abies grandis
|
| 73 | c3dzoA_ |
|
not modelled |
7.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:rhoptry kinase domain;
PDBTitle: crystal structure of a rhoptry kinase from toxoplasma gondii
|
| 74 | c3dfgA_ |
|
not modelled |
6.6 |
15 |
PDB header:recombination
Chain: A: PDB Molecule:regulatory protein recx;
PDBTitle: crystal structure of recx: a potent inhibitor protein of2 reca from xanthomonas campestris
|
| 75 | c2jiiA_ |
|
not modelled |
6.3 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine-protein kinase vrk3 molecule: vaccinia
PDBTitle: structure of vaccinia related kinase 3
|
| 76 | c3pjaK_ |
|
not modelled |
6.3 |
13 |
PDB header:hydrolase
Chain: K: PDB Molecule:translin-associated protein x;
PDBTitle: crystal structure of human c3po complex
|
| 77 | d2fswa1 |
|
not modelled |
6.1 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:HxlR-like |
| 78 | c2j5cB_ |
|
not modelled |
5.4 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:1,8-cineole synthase;
PDBTitle: rational conversion of substrate and product specificity2 in a monoterpene synthase. structural insights into the3 molecular basis of rapid evolution.
|
| 79 | d1s2xa_ |
|
not modelled |
5.3 |
50 |
Fold:STAT-like
Superfamily:Cag-Z
Family:Cag-Z |
| 80 | c1s2xA_ |
|
not modelled |
5.3 |
50 |
PDB header:unknown function
Chain: A: PDB Molecule:cag-z;
PDBTitle: crystal structure of cag-z from helicobacter pylori
|
| 81 | c2pziA_ |
|
not modelled |
5.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:probable serine/threonine-protein kinase pkng;
PDBTitle: crystal structure of protein kinase pkng from mycobacterium2 tuberculosis in complex with tetrahydrobenzothiophene ax20017
|