| 1 | c2w48D_
|
|
 |
100.0 |
22 |
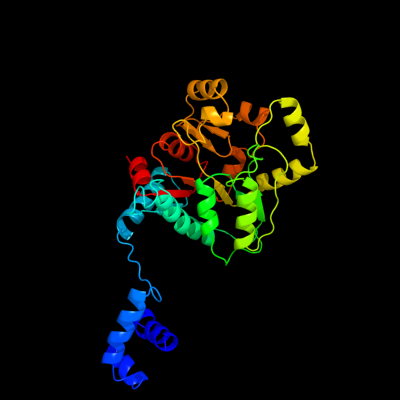
PDB header:transcription
Chain: D: PDB Molecule:sorbitol operon regulator;
PDBTitle: crystal structure of the full-length sorbitol operon2 regulator sorc from klebsiella pneumoniae
|
| 2 | c3kv1A_
|
|
 |
100.0 |
22 |
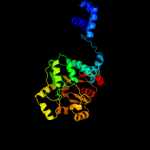
PDB header:transcription
Chain: A: PDB Molecule:transcriptional repressor;
PDBTitle: crystal structure of putative sugar-binding domain of transcriptional2 repressor from vibrio fischeri
|
| 3 | c3nzeB_
|
|
 |
100.0 |
22 |
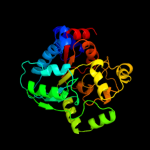
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator, sugar-binding family;
PDBTitle: the crystal structure of a domain of a possible sugar-binding2 transcriptional regulator from arthrobacter aurescens tc1.
|
| 4 | d2gnpa1
|
|
 |
100.0 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 5 | d3efba1
|
|
 |
100.0 |
22 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 6 | d2okga1
|
|
 |
100.0 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 7 | d2o0ma1
|
|
 |
100.0 |
23 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 8 | c2o0mA_
|
|
 |
100.0 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, sorc family;
PDBTitle: the crystal structure of the putative sorc family transcriptional2 regulator from enterococcus faecalis
|
| 9 | d2r5fa1
|
|
 |
100.0 |
32 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 10 | c2bkxB_
|
|
 |
100.0 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: structure and kinetics of a monomeric glucosamine-6-2 phosphate deaminase: missing link of the nagb superfamily
|
| 11 | c2ri0B_
|
|
 |
100.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine 6-phosphate deaminase (nagb) from s.2 mutans
|
| 12 | d1ne7a_
|
|
 |
99.5 |
17 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
| 13 | c3hn6D_
|
|
 |
99.4 |
14 |
PDB header:isomerase
Chain: D: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine-6-phosphate deaminase from borrelia2 burgdorferi
|
| 14 | d1fsfa_
|
|
 |
99.3 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
| 15 | c3e15D_
|
|
 |
99.3 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: 6-phosphogluconolactonase from plasmodium vivax
|
| 16 | c3nwpA_
|
|
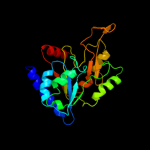 |
99.1 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of a 6-phosphogluconolactonase (sbal_2240) from2 shewanella baltica os155 at 1.40 a resolution
|
| 17 | c3icoA_
|
|
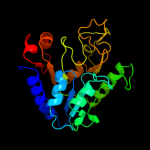 |
99.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from2 mycobacterium tuberculosis
|
| 18 | c3oc6A_
|
|
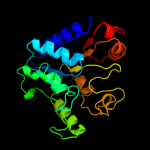 |
99.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from mycobacterium2 smegmatis, apo form
|
| 19 | c3lhiA_
|
|
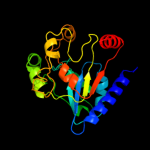 |
99.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative 6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-2 phosphogluconolactonase(yp_207848.1) from neisseria3 gonorrhoeae fa 1090 at 1.33 a resolution
|
| 20 | d1vl1a_
|
|
 |
99.0 |
12 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
| 21 | c3lwdA_ |
|
not modelled |
99.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-phosphogluconolactonase (yp_574786.1)2 from chromohalobacter salexigens dsm 3043 at 1.88 a resolution
|
| 22 | c2j0eA_ |
|
not modelled |
99.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: three dimensional structure and catalytic mechanism of 6-2 phosphogluconolactonase from trypanosoma brucei
|
| 23 | c1y89B_ |
|
not modelled |
99.0 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:devb protein;
PDBTitle: crystal structure of devb protein
|
| 24 | c3cssA_ |
|
not modelled |
99.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from leishmania2 guyanensis
|
| 25 | c1pbtA_ |
|
not modelled |
99.0 |
11 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: the crystal structure of tm1154, oxidoreductase, sol/devb2 family from thermotoga maritima
|
| 26 | c3u7jA_ |
|
not modelled |
97.9 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
|
| 27 | c1rp3G_ |
|
not modelled |
97.5 |
27 |
PDB header:transcription
Chain: G: PDB Molecule:rna polymerase sigma factor sigma-28 (flia);
PDBTitle: cocrystal structure of the flagellar sigma/anti-sigma2 complex, sigma-28/flgm
|
| 28 | c3ctaA_ |
|
not modelled |
97.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin kinase;
PDBTitle: crystal structure of riboflavin kinase from thermoplasma2 acidophilum
|
| 29 | d1i5za1 |
|
not modelled |
97.5 |
27 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 30 | d2bgca1 |
|
not modelled |
97.5 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 31 | d1s7oa_ |
|
not modelled |
97.4 |
31 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:YlxM/p13-like |
| 32 | d2oz6a1 |
|
not modelled |
97.4 |
36 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 33 | c3kwmC_ |
|
not modelled |
97.4 |
22 |
PDB header:isomerase
Chain: C: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-isomerase a
|
| 34 | c3d0sA_ |
|
not modelled |
97.3 |
32 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein;
PDBTitle: camp receptor protein from m.tuberculosis, camp-free form
|
| 35 | d1xsva_ |
|
not modelled |
97.3 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:YlxM/p13-like |
| 36 | d2gaua1 |
|
not modelled |
97.3 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 37 | d2h6ca1 |
|
not modelled |
97.2 |
26 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 38 | c3kccA_ |
|
not modelled |
97.2 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:catabolite gene activator;
PDBTitle: crystal structure of d138l mutant of catabolite gene activator protein
|
| 39 | d1k78a1 |
|
not modelled |
97.2 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Paired domain |
| 40 | c2gauA_ |
|
not modelled |
97.2 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, crp/fnr family;
PDBTitle: crystal structure of transcriptional regulator, crp/fnr family from2 porphyromonas gingivalis (apc80792), structural genomics, mcsg
|
| 41 | c2oz6A_ |
|
not modelled |
97.2 |
36 |
PDB header:dna binding protein
Chain: A: PDB Molecule:virulence factor regulator;
PDBTitle: crystal structure of virulence factor regulator from pseudomonas2 aeruginosa in complex with camp
|
| 42 | d2coha1 |
|
not modelled |
97.2 |
27 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 43 | c1zreB_ |
|
not modelled |
97.1 |
27 |
PDB header:gene regulation/dna
Chain: B: PDB Molecule:catabolite gene activator;
PDBTitle: 4 crystal structures of cap-dna with all base-pair2 substitutions at position 6, cap-[6g;17c]icap38 dna
|
| 44 | d6paxa1 |
|
not modelled |
97.1 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Paired domain |
| 45 | d2d1ha1 |
|
not modelled |
97.1 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:TrmB-like |
| 46 | d1ft9a1 |
|
not modelled |
97.1 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 47 | c2bgcA_ |
|
not modelled |
97.1 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:prfa;
PDBTitle: prfa-g145s, a constitutive active mutant of the2 transcriptional regulator in l.monocytogenes
|
| 48 | c6paxA_ |
|
not modelled |
97.1 |
22 |
PDB header:gene regulation/dna
Chain: A: PDB Molecule:homeobox protein pax-6;
PDBTitle: crystal structure of the human pax-6 paired domain-dna2 complex reveals a general model for pax protein-dna3 interactions
|
| 49 | c3dv8A_ |
|
not modelled |
97.1 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, crp/fnr family;
PDBTitle: crystal structure of a putative transcriptional regulator of the2 crp/fnr family (eubrec_1222) from eubacterium rectale atcc 33656 at3 2.55 a resolution
|
| 50 | d3e5ua1 |
|
not modelled |
97.1 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CAP C-terminal domain-like |
| 51 | c3e6dA_ |
|
not modelled |
97.1 |
26 |
PDB header:transcription regulation
Chain: A: PDB Molecule:cyclic nucleotide-binding protein;
PDBTitle: crystal structure of cprk c200s
|
| 52 | c1u78A_ |
|
not modelled |
97.0 |
18 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:transposable element tc3 transposase;
PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
|
| 53 | c2o8xA_ |
|
not modelled |
97.0 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:probable rna polymerase sigma-c factor;
PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
|
| 54 | d1pdnc_ |
|
not modelled |
97.0 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Paired domain |
| 55 | c1ft9A_ |
|
not modelled |
97.0 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:carbon monoxide oxidation system transcription
PDBTitle: structure of the reduced (feii) co-sensing protein from r.2 rubrum
|
| 56 | c3hheA_ |
|
not modelled |
97.0 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from bartonella2 henselae
|
| 57 | c1zybA_ |
|
not modelled |
97.0 |
21 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcription regulator, crp family;
PDBTitle: crystal structure of transcription regulator from bacteroides2 thetaiotaomicron vpi-5482 at 2.15 a resolution
|
| 58 | d1ku7a_ |
|
not modelled |
97.0 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 59 | d1sfxa_ |
|
not modelled |
97.0 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:TrmB-like |
| 60 | d1z05a1 |
|
not modelled |
97.0 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ROK associated domain |
| 61 | c3hugA_ |
|
not modelled |
96.9 |
25 |
PDB header:transcription/membrane protein
Chain: A: PDB Molecule:rna polymerase sigma factor;
PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
|
| 62 | c2zcwA_ |
|
not modelled |
96.9 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, fnr/crp family;
PDBTitle: crystal structure of ttha1359, a transcriptional regulator,2 crp/fnr family from thermus thermophilus hb8
|
| 63 | c3tgnA_ |
|
not modelled |
96.9 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:adc operon repressor adcr;
PDBTitle: crystal structure of the zinc-dependent marr family transcriptional2 regulator adcr in the zn(ii)-bound state
|
| 64 | c2qwwB_ |
|
not modelled |
96.9 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
|
| 65 | d1rp3a2 |
|
not modelled |
96.9 |
27 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 66 | c3iwzB_ |
|
not modelled |
96.9 |
29 |
PDB header:transcription
Chain: B: PDB Molecule:catabolite activation-like protein;
PDBTitle: the c-di-gmp responsive global regulator clp links cell-cell signaling2 to virulence gene expression in xanthomonas campestris
|
| 67 | c1or7A_ |
|
not modelled |
96.9 |
39 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma-e factor;
PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
|
| 68 | c3g3zA_ |
|
not modelled |
96.9 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
|
| 69 | d1ku9a_ |
|
not modelled |
96.9 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DNA-binding protein Mj223 |
| 70 | c1zx4B_ |
|
not modelled |
96.9 |
33 |
PDB header:translation
Chain: B: PDB Molecule:plasmid partition par b protein;
PDBTitle: structure of parb bound to dna
|
| 71 | c3fx3A_ |
|
not modelled |
96.9 |
11 |
PDB header:camp-binding protein
Chain: A: PDB Molecule:cyclic nucleotide-binding protein;
PDBTitle: structure of a putative camp-binding regulatory protein from2 silicibacter pomeroyi dss-3
|
| 72 | c3mzyA_ |
|
not modelled |
96.9 |
22 |
PDB header:rna binding protein
Chain: A: PDB Molecule:rna polymerase sigma-h factor;
PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
|
| 73 | c2it0A_ |
|
not modelled |
96.8 |
14 |
PDB header:transcription/dna
Chain: A: PDB Molecule:iron-dependent repressor ider;
PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
|
| 74 | c2f8mB_ |
|
not modelled |
96.8 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: ribose 5-phosphate isomerase from plasmodium falciparum
|
| 75 | c2wteB_ |
|
not modelled |
96.8 |
19 |
PDB header:antiviral protein
Chain: B: PDB Molecule:csa3;
PDBTitle: the structure of the crispr-associated protein, csa3, from2 sulfolobus solfataricus at 1.8 angstrom resolution.
|
| 76 | c3la2A_ |
|
not modelled |
96.8 |
32 |
PDB header:transcription
Chain: A: PDB Molecule:global nitrogen regulator;
PDBTitle: crystal structure of ntca in complex with 2-oxoglutarate
|
| 77 | d1ttya_ |
|
not modelled |
96.8 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 78 | d2fxaa1 |
|
not modelled |
96.8 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 79 | c2fxaB_ |
|
not modelled |
96.8 |
19 |
PDB header:transcription
Chain: B: PDB Molecule:protease production regulatory protein hpr;
PDBTitle: structure of the protease production regulatory protein hpr from2 bacillus subtilis.
|
| 80 | c2h09A_ |
|
not modelled |
96.7 |
28 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator mntr;
PDBTitle: crystal structure of diphtheria toxin repressor like protein2 from e. coli
|
| 81 | c1f5tA_ |
|
not modelled |
96.7 |
16 |
PDB header:transcription/dna
Chain: A: PDB Molecule:diphtheria toxin repressor;
PDBTitle: diphtheria tox repressor (c102d mutant) complexed with2 nickel and dtxr consensus binding sequence
|
| 82 | c1m0sA_ |
|
not modelled |
96.7 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: northeast structural genomics consortium (nesg id ir21)
|
| 83 | d3ctaa1 |
|
not modelled |
96.7 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 84 | d1or7a1 |
|
not modelled |
96.7 |
38 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 85 | d2etha1 |
|
not modelled |
96.7 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 86 | c3e97A_ |
|
not modelled |
96.7 |
18 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, crp/fnr family;
PDBTitle: crystal structure of transcriptional regulator of crp/fnr2 family (yp_604437.1) from deinococcus geothermalis dsm3 11300 at 1.86 a resolution
|
| 87 | c3nrvC_ |
|
not modelled |
96.7 |
14 |
PDB header:transcription regulator
Chain: C: PDB Molecule:putative transcriptional regulator (marr/emrr family);
PDBTitle: crystal structure of marr/emrr family transcriptional regulator from2 acinetobacter sp. adp1
|
| 88 | c3bddD_ |
|
not modelled |
96.7 |
18 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein marr;
PDBTitle: crystal structure of a putative multiple antibiotic-resistance2 repressor (ssu05_1136) from streptococcus suis 89/1591 at 2.20 a3 resolution
|
| 89 | c1lkzB_ |
|
not modelled |
96.7 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase a;
PDBTitle: crystal structure of d-ribose-5-phosphate isomerase (rpia)2 from escherichia coli.
|
| 90 | c1z05A_ |
|
not modelled |
96.6 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, rok family;
PDBTitle: crystal structure of the rok family transcriptional regulator, homolog2 of e.coli mlc protein.
|
| 91 | c1lk5C_ |
|
not modelled |
96.6 |
23 |
PDB header:isomerase
Chain: C: PDB Molecule:d-ribose-5-phosphate isomerase;
PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
|
| 92 | d1ku3a_ |
|
not modelled |
96.6 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 93 | c2nnnB_ |
|
not modelled |
96.6 |
13 |
PDB header:transcription
Chain: B: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of probable transcriptional regulator from2 pseudomonas aeruginosa
|
| 94 | d1z6ra1 |
|
not modelled |
96.6 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ROK associated domain |
| 95 | c3nqoB_ |
|
not modelled |
96.6 |
14 |
PDB header:transcription
Chain: B: PDB Molecule:marr-family transcriptional regulator;
PDBTitle: crystal structure of a marr family transcriptional regulator (cd1569)2 from clostridium difficile 630 at 2.20 a resolution
|
| 96 | c3deuB_ |
|
not modelled |
96.6 |
20 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator slya;
PDBTitle: crystal structure of transcription regulatory protein slya2 from salmonella typhimurium in complex with salicylate3 ligands
|
| 97 | c3r0aB_ |
|
not modelled |
96.6 |
12 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator;
PDBTitle: possible transcriptional regulator from methanosarcina mazei go1 (gi2 21227196)
|
| 98 | c2fa5B_ |
|
not modelled |
96.6 |
25 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator marr/emrr family;
PDBTitle: the crystal structure of an unliganded multiple antibiotic-2 resistance repressor (marr) from xanthomonas campestris
|
| 99 | d1lj9a_ |
|
not modelled |
96.6 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 100 | d3broa1 |
|
not modelled |
96.6 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 101 | c2zdbA_ |
|
not modelled |
96.5 |
30 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, crp family;
PDBTitle: crystal structure of tthb099, a transcriptional regulator crp family2 from thermus thermophilus hb8
|
| 102 | c2fmyB_ |
|
not modelled |
96.5 |
28 |
PDB header:dna binding protein
Chain: B: PDB Molecule:carbon monoxide oxidation system transcription regulator
PDBTitle: co-dependent transcription factor cooa from carboxydothermus2 hydrogenoformans (imidazole-bound form)
|
| 103 | c2nyxB_ |
|
not modelled |
96.5 |
10 |
PDB header:transcription
Chain: B: PDB Molecule:probable transcriptional regulatory protein, rv1404;
PDBTitle: crystal structure of rv1404 from mycobacterium tuberculosis
|
| 104 | c2ev5B_ |
|
not modelled |
96.5 |
15 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator mntr;
PDBTitle: bacillus subtilis manganese transport regulator (mntr)2 bound to calcium
|
| 105 | c3bpxB_ |
|
not modelled |
96.5 |
21 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of marr
|
| 106 | c3dkwB_ |
|
not modelled |
96.5 |
16 |
PDB header:transcription regulator
Chain: B: PDB Molecule:dnr protein;
PDBTitle: crystal structure of dnr from pseudomonas aeruginosa.
|
| 107 | c2l4aA_
|
|
 |
96.5 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:leucine responsive regulatory protein;
PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
|
| 108 | c2x4hA_ |
|
not modelled |
96.4 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:hypothetical protein sso2273;
PDBTitle: crystal structure of the hypothetical protein sso2273 from2 sulfolobus solfataricus
|
| 109 | c1z6rC_ |
|
not modelled |
96.4 |
14 |
PDB header:transcription
Chain: C: PDB Molecule:mlc protein;
PDBTitle: crystal structure of mlc from escherichia coli
|
| 110 | c2rdpA_ |
|
not modelled |
96.4 |
20 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator marr;
PDBTitle: the structure of a marr family protein from bacillus2 stearothermophilus
|
| 111 | c3k0lA_ |
|
not modelled |
96.4 |
19 |
PDB header:transcription regulator
Chain: A: PDB Molecule:repressor protein;
PDBTitle: crystal structure of putative marr family transcriptional2 regulator from acinetobacter sp. adp
|
| 112 | c3l7oB_ |
|
not modelled |
96.4 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
| 113 | c2pexA_ |
|
not modelled |
96.4 |
22 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator ohrr;
PDBTitle: structure of reduced c22s ohrr from xanthamonas campestris
|
| 114 | d1lnwa_ |
|
not modelled |
96.3 |
8 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 115 | d2p7vb1 |
|
not modelled |
96.3 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 116 | d2hr3a1 |
|
not modelled |
96.3 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 117 | c2gxgA_ |
|
not modelled |
96.3 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:146aa long hypothetical transcriptional regulator;
PDBTitle: crystal structure of emrr homolog from hyperthermophilic archaea2 sulfolobus tokodaii strain7
|
| 118 | d1s3ja_ |
|
not modelled |
96.3 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 119 | c2e7xA_ |
|
not modelled |
96.3 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:150aa long hypothetical transcriptional regulator;
PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
|
| 120 | d2fbha1 |
|
not modelled |
96.3 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |