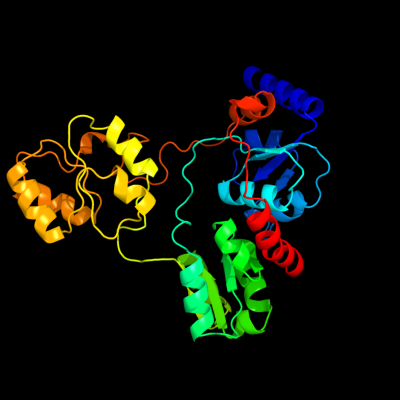
| 1 | c2qs0A_
|
|
 |
100.0 |
39 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:quinolinate synthetase a;
PDBTitle: quinolinate synthase from pyrococcus furiosus
|
| 2 | d1wzua1
|
|
 |
100.0 |
40 |
Fold:NadA-like
Superfamily:NadA-like
Family:NadA-like |
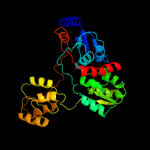
| 3 | c3dmpD_
|
|
 |
74.2 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: 2.6 a crystal structure of uracil phosphoribosyltransferase2 from burkholderia pseudomallei
|
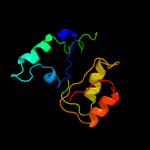
| 4 | d2nx2a1
|
|
 |
70.1 |
10 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:YpsA-like |
| 5 | c2e55D_
|
|
 |
61.9 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: structure of aq2163 protein from aquifex aeolicus
|
| 6 | c2ekcA_
|
|
 |
59.5 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 7 | c3fn9B_
|
|
 |
57.9 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
| 8 | c3cmgA_
|
|
 |
55.2 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
| 9 | d16pka_
|
|
 |
52.6 |
21 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 10 | d1xhja_
|
|
 |
51.4 |
12 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:NifU C-terminal domain-like |
| 11 | c3gm8A_
|
|
 |
48.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycoside hydrolase family 2, candidate beta-glycosidase;
PDBTitle: crystal structure of a beta-glycosidase from bacteroides vulgatus
|
| 12 | d2tpsa_
|
|
 |
47.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 13 | d1j97a_
|
|
 |
46.8 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:Phosphoserine phosphatase |
| 14 | d1jbqa_
|
|
 |
46.7 |
20 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 15 | c1jbqD_
|
|
 |
46.7 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:cystathionine beta-synthase;
PDBTitle: structure of human cystathionine beta-synthase: a unique pyridoxal 5'-2 phosphate dependent hemeprotein
|
| 16 | d1ybha1
|
|
 |
45.6 |
20 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 17 | c3iz6A_
|
|
 |
45.1 |
15 |
PDB header:ribosome
Chain: A: PDB Molecule:40s ribosomal protein sa (s2p);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 18 | c3k7pA_
|
|
 |
42.0 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
|
| 19 | d1vhca_
|
|
 |
41.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 20 | d2ez9a1
|
|
 |
40.1 |
18 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 21 | c3m1pA_ |
|
not modelled |
39.5 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
|
| 22 | c3p04B_ |
|
not modelled |
39.4 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized bcr;
PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
|
| 23 | d1ojra_ |
|
not modelled |
38.7 |
7 |
Fold:AraD/HMP-PK domain-like
Superfamily:AraD/HMP-PK domain-like
Family:AraD-like aldolase/epimerase |
| 24 | c2pcjB_ |
|
not modelled |
38.5 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:lipoprotein-releasing system atp-binding protein lold;
PDBTitle: crystal structure of abc transporter (aq_297) from aquifex aeolicus2 vf5
|
| 25 | c3p9zA_ |
|
not modelled |
37.6 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
| 26 | d1jr2a_ |
|
not modelled |
37.4 |
19 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
| 27 | c1jr2A_ |
|
not modelled |
37.4 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: structure of uroporphyrinogen iii synthase
|
| 28 | c1s1hB_ |
|
not modelled |
37.3 |
18 |
PDB header:ribosome
Chain: B: PDB Molecule:40s ribosomal protein s0-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 29 | c2ejaB_ |
|
not modelled |
33.3 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
| 30 | c1zq1B_ |
|
not modelled |
33.2 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
|
| 31 | d1zbra1 |
|
not modelled |
32.5 |
20 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
| 32 | c3p04A_ |
|
not modelled |
32.1 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized bcr;
PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
|
| 33 | c2infB_ |
|
not modelled |
31.7 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 bacillus subtilis
|
| 34 | c3bg3B_ |
|
not modelled |
31.3 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
|
| 35 | d1hrua_ |
|
not modelled |
29.8 |
15 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:YrdC-like |
| 36 | c1d7kB_ |
|
not modelled |
29.7 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:human ornithine decarboxylase;
PDBTitle: crystal structure of human ornithine decarboxylase at 2.12 angstroms resolution
|
| 37 | d1veha_ |
|
not modelled |
29.4 |
9 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:NifU C-terminal domain-like |
| 38 | d1zq1a2 |
|
not modelled |
29.3 |
26 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 39 | c1oidA_ |
|
not modelled |
28.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein usha;
PDBTitle: 5'-nucleotidase (e. coli) with an engineered disulfide2 bridge (s228c, p513c)
|
| 40 | d1bd3a_ |
|
not modelled |
28.6 |
21 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 41 | d2awna2 |
|
not modelled |
28.5 |
23 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 42 | d1bw0a_ |
|
not modelled |
28.2 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 43 | c2p5tA_ |
|
not modelled |
27.6 |
30 |
PDB header:transcription regulator
Chain: A: PDB Molecule:putative transcriptional regulator peza;
PDBTitle: molecular and structural characterization of the pezat chromosomal2 toxin-antitoxin system of the human pathogen streptococcus pneumoniae
|
| 44 | d2pjua1 |
|
not modelled |
27.5 |
17 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 45 | c3dwgA_ |
|
not modelled |
26.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase b;
PDBTitle: crystal structure of a sulfur carrier protein complex found in the2 cysteine biosynthetic pathway of mycobacterium tuberculosis
|
| 46 | d1agxa_ |
|
not modelled |
26.4 |
29 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 47 | d1xkna_ |
|
not modelled |
26.2 |
16 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
| 48 | d1pvda1 |
|
not modelled |
26.2 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 49 | d1wbha1 |
|
not modelled |
25.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 50 | d1v9sa1 |
|
not modelled |
25.1 |
24 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 51 | d1pswa_ |
|
not modelled |
25.0 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:ADP-heptose LPS heptosyltransferase II |
| 52 | c2xznB_ |
|
not modelled |
24.8 |
19 |
PDB header:ribosome
Chain: B: PDB Molecule:rps0e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 53 | d1b0ua_ |
|
not modelled |
24.2 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 54 | d1phpa_ |
|
not modelled |
24.1 |
17 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 55 | c3hbmA_ |
|
not modelled |
23.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:udp-sugar hydrolase;
PDBTitle: crystal structure of pseg from campylobacter jejuni
|
| 56 | c3nm3D_ |
|
not modelled |
23.3 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:thiamine biosynthetic bifunctional enzyme;
PDBTitle: the crystal structure of candida glabrata thi6, a bifunctional enzyme2 involved in thiamin biosyhthesis of eukaryotes
|
| 57 | d1jcua_ |
|
not modelled |
22.9 |
10 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:YrdC-like |
| 58 | c2yswB_ |
|
not modelled |
22.8 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of the 3-dehydroquinate dehydratase from aquifex2 aeolicus vf5
|
| 59 | c2htmB_ |
|
not modelled |
22.8 |
23 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiazole biosynthesis protein thig;
PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
|
| 60 | d1g2912 |
|
not modelled |
22.0 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 61 | c2ehhE_ |
|
not modelled |
20.3 |
19 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 62 | c3tovB_ |
|
not modelled |
20.0 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase family 9;
PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
|
| 63 | c3bchA_ |
|
not modelled |
19.9 |
14 |
PDB header:cell adhesion, ribosomal protein
Chain: A: PDB Molecule:40s ribosomal protein sa;
PDBTitle: crystal structure of the human laminin receptor precursor
|
| 64 | d1i5ea_ |
|
not modelled |
19.7 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 65 | c3bgaB_ |
|
not modelled |
19.3 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of beta-galactosidase from bacteroides2 thetaiotaomicron vpi-5482
|
| 66 | c3il0B_ |
|
not modelled |
17.7 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:aminopeptidase p; xaa-pro aminopeptidase;
PDBTitle: the crystal structure of the aminopeptidase p,xaa-pro aminopeptidase2 from streptococcus thermophilus
|
| 67 | c2yz2B_ |
|
not modelled |
17.5 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative abc transporter atp-binding protein tm_0222;
PDBTitle: crystal structure of the abc transporter in the cobalt transport2 system
|
| 68 | d1usha2 |
|
not modelled |
17.4 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 69 | c3jyfB_ |
|
not modelled |
17.2 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-
PDBTitle: the crystal structure of a 2,3-cyclic nucleotide 2-2 phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor3 protein from klebsiella pneumoniae subsp. pneumoniae mgh 78578
|
| 70 | c2jnvA_ |
|
not modelled |
16.8 |
14 |
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 1, chloroplast;
PDBTitle: solution structure of c-terminal domain of nifu-like2 protein from oryza sativa
|
| 71 | d3dhwc1 |
|
not modelled |
16.7 |
7 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 72 | d2d6fa2 |
|
not modelled |
16.4 |
24 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 73 | c3nxkE_ |
|
not modelled |
16.3 |
29 |
PDB header:hydrolase
Chain: E: PDB Molecule:cytoplasmic l-asparaginase;
PDBTitle: crystal structure of probable cytoplasmic l-asparaginase from2 campylobacter jejuni
|
| 74 | c1zmrA_ |
|
not modelled |
16.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of the e. coli phosphoglycerate kinase
|
| 75 | d1r3sa_ |
|
not modelled |
16.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
| 76 | d2ocda1 |
|
not modelled |
16.1 |
24 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 77 | d2vzsa5 |
|
not modelled |
16.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 78 | c2k4mA_ |
|
not modelled |
16.0 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0146 protein mth_1000;
PDBTitle: solution nmr structure of m. thermoautotrophicum protein2 mth_1000, northeast structural genomics consortium target3 tr8
|
| 79 | c3pihA_ |
|
not modelled |
16.0 |
20 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:uvrabc system protein a;
PDBTitle: t. maritima uvra in complex with fluorescein-modified dna
|
| 80 | c1zfjA_ |
|
not modelled |
15.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 81 | c2e0kA_ |
|
not modelled |
15.6 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 c20-methyltransferase;
PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
|
| 82 | c2vzvB_ |
|
not modelled |
15.6 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:exo-beta-d-glucosaminidase;
PDBTitle: substrate complex of amycolatopsis orientalis exo-2 chitosanase csxa e541a with chitosan
|
| 83 | c1q1bD_ |
|
not modelled |
15.5 |
21 |
PDB header:transport protein
Chain: D: PDB Molecule:maltose/maltodextrin transport atp-binding protein malk;
PDBTitle: crystal structure of e. coli malk in the nucleotide-free form
|
| 84 | c2r8zC_ |
|
not modelled |
15.4 |
30 |
PDB header:hydrolase
Chain: C: PDB Molecule:3-deoxy-d-manno-octulosonate 8-phosphate phosphatase;
PDBTitle: crystal structure of yrbi phosphatase from escherichia coli in complex2 with a phosphate and a calcium ion
|
| 85 | d1pvta_ |
|
not modelled |
15.4 |
10 |
Fold:AraD/HMP-PK domain-like
Superfamily:AraD/HMP-PK domain-like
Family:AraD-like aldolase/epimerase |
| 86 | c2i4rA_ |
|
not modelled |
15.3 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:v-type atp synthase subunit f;
PDBTitle: crystal structure of the v-type atp synthase subunit f from2 archaeoglobus fulgidus. nesg target gr52a.
|
| 87 | c2kg4A_ |
|
not modelled |
15.2 |
32 |
PDB header:cell cycle
Chain: A: PDB Molecule:growth arrest and dna-damage-inducible protein
PDBTitle: three-dimensional structure of human gadd45alpha in2 solution by nmr
|
| 88 | c1wnfA_ |
|
not modelled |
15.2 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of ph0066 from pyrococcus horikoshii
|
| 89 | c2d3wB_ |
|
not modelled |
15.1 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:probable atp-dependent transporter sufc;
PDBTitle: crystal structure of escherichia coli sufc, an atpase2 compenent of the suf iron-sulfur cluster assembly machinery
|
| 90 | c3m1yA_ |
|
not modelled |
15.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase (serb);
PDBTitle: crystal structure of a phosphoserine phosphatase (serb) from2 helicobacter pylori
|
| 91 | c1nm3B_ |
|
not modelled |
14.7 |
24 |
PDB header:electron transport
Chain: B: PDB Molecule:protein hi0572;
PDBTitle: crystal structure of heamophilus influenza hybrid-prx5
|
| 92 | c2yxgD_ |
|
not modelled |
14.7 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 93 | c2olkD_ |
|
not modelled |
14.7 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:amino acid abc transporter;
PDBTitle: abc protein artp in complex with adp-beta-s
|
| 94 | c2o7qA_ |
|
not modelled |
14.7 |
15 |
PDB header:oxidoreductase,transferase
Chain: A: PDB Molecule:bifunctional 3-dehydroquinate dehydratase/shikimate
PDBTitle: crystal structure of the a. thaliana dhq-dehydroshikimate-sdh-2 shikimate-nadp(h)
|
| 95 | d1ji0a_ |
|
not modelled |
14.7 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 96 | d1g6ha_ |
|
not modelled |
14.6 |
9 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 97 | c3eebB_ |
|
not modelled |
14.6 |
25 |
PDB header:toxin
Chain: B: PDB Molecule:rtx toxin rtxa;
PDBTitle: structure of the v. cholerae rtx cysteine protease domain
|
| 98 | c3mc3A_ |
|
not modelled |
14.5 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsre/dsrf-like family protein;
PDBTitle: crystal structure of dsre/dsrf-like family protein (np_342589.1) from2 sulfolobus solfataricus at 1.49 a resolution
|
| 99 | d1wsaa_ |
|
not modelled |
14.3 |
24 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |