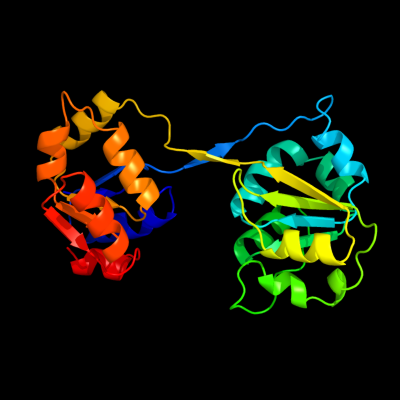
| 1 | c3re1B_
|
|
 |
100.0 |
27 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthetase;
PDBTitle: crystal structure of uroporphyrinogen iii synthase from pseudomonas2 syringae pv. tomato dc3000
|
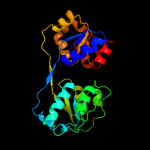
| 2 | c1jr2A_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: structure of uroporphyrinogen iii synthase
|
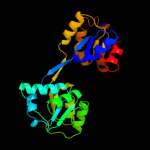
| 3 | d1jr2a_
|
|
 |
100.0 |
17 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
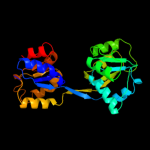
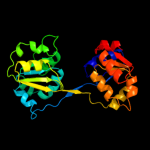
| 4 | c3mw8A_
|
|
 |
100.0 |
37 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of an uroporphyrinogen-iii synthase (sama_3255) from2 shewanella amazonensis sb2b at 1.65 a resolution
|
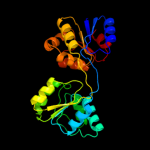
| 5 | c3d8tB_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
|
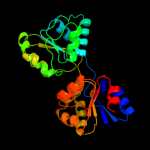
| 6 | d1wd7a_
|
|
 |
100.0 |
18 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
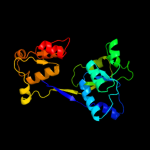
| 7 | c3p9zA_
|
|
 |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
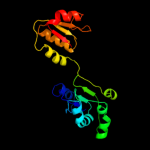
| 8 | c1ybaC_
|
|
 |
96.1 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: the active form of phosphoglycerate dehydrogenase
|
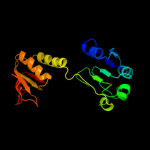
| 9 | c3k5pA_
|
|
 |
95.5 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
|
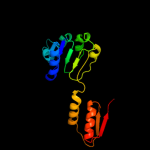
| 10 | c2eklA_
|
|
 |
95.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: structure of st1218 protein from sulfolobus tokodaii
|
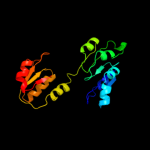
| 11 | c2cukC_
|
|
 |
94.8 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
| 12 | d1rtta_
|
|
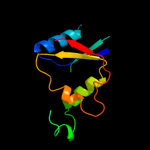 |
92.8 |
10 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 13 | c2gcgB_
|
|
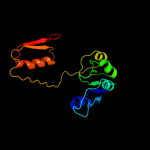 |
92.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
| 14 | c1wwkA_
|
|
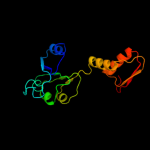 |
91.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
| 15 | c2o4cB_
|
|
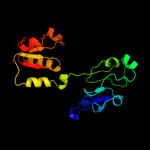 |
88.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:erythronate-4-phosphate dehydrogenase;
PDBTitle: crystal structure of d-erythronate-4-phosphate dehydrogenase complexed2 with nad
|
| 16 | d7reqa2
|
|
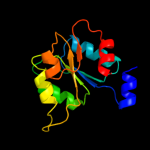 |
87.9 |
18 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 17 | c2yxbA_
|
|
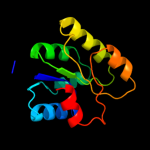 |
87.9 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 18 | c1xdwA_
|
|
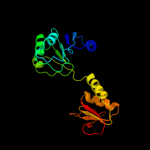 |
86.7 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad+-dependent (r)-2-hydroxyglutarate
PDBTitle: nad+-dependent (r)-2-hydroxyglutarate dehydrogenase from2 acidaminococcus fermentans
|
| 19 | c1dxyA_
|
|
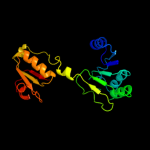 |
86.5 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-2-hydroxyisocaproate dehydrogenase;
PDBTitle: structure of d-2-hydroxyisocaproate dehydrogenase
|
| 20 | c3ksmA_
|
|
 |
86.5 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
| 21 | d2qwxa1 |
|
not modelled |
85.8 |
28 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 22 | d1lssa_ |
|
not modelled |
83.2 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 23 | c3hn0A_ |
|
not modelled |
82.8 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein;
PDBTitle: crystal structure of an abc transporter (bdi_1369) from2 parabacteroides distasonis at 1.75 a resolution
|
| 24 | c3l7nA_ |
|
not modelled |
82.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of smu.1228c
|
| 25 | c1lk5C_ |
|
not modelled |
80.7 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:d-ribose-5-phosphate isomerase;
PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
|
| 26 | c3eywA_ |
|
not modelled |
80.1 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 27 | c2g76A_ |
|
not modelled |
79.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 28 | c3lcmB_ |
|
not modelled |
79.8 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
|
| 29 | d1ccwa_ |
|
not modelled |
79.0 |
14 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 30 | c3o1hB_ |
|
not modelled |
78.5 |
12 |
PDB header:signaling protein
Chain: B: PDB Molecule:periplasmic protein tort;
PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
|
| 31 | c1y80A_ |
|
not modelled |
78.4 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 32 | c3e61A_ |
|
not modelled |
77.9 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative transcriptional repressor of ribose operon;
PDBTitle: crystal structure of a putative transcriptional repressor of ribose2 operon from staphylococcus saprophyticus subsp. saprophyticus
|
| 33 | c2fzvC_ |
|
not modelled |
77.9 |
20 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative arsenical resistance protein;
PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
|
| 34 | d1ydga_ |
|
not modelled |
77.8 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 35 | c1k5hB_ |
|
not modelled |
77.8 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose-5-phosphate reductoisomerase;
PDBTitle: 1-deoxy-d-xylulose-5-phosphate reductoisomerase
|
| 36 | d2fzva1 |
|
not modelled |
77.6 |
23 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 37 | d1dxqa_ |
|
not modelled |
77.6 |
30 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 38 | c3orqA_ |
|
not modelled |
76.8 |
12 |
PDB header:ligase,biosynthetic protein
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide synthetase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
|
| 39 | d1sc6a2 |
|
not modelled |
76.3 |
12 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 40 | c2pjuD_ |
|
not modelled |
75.9 |
13 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 41 | d1o1ya_ |
|
not modelled |
75.7 |
14 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 42 | c3pdiB_ |
|
not modelled |
74.7 |
15 |
PDB header:protein binding
Chain: B: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nifn;
PDBTitle: precursor bound nifen
|
| 43 | c2f8mB_ |
|
not modelled |
74.4 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: ribose 5-phosphate isomerase from plasmodium falciparum
|
| 44 | c3q2oB_ |
|
not modelled |
74.2 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase, atpase subunit;
PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
|
| 45 | c2qk4A_ |
|
not modelled |
74.0 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:trifunctional purine biosynthetic protein adenosine-3;
PDBTitle: human glycinamide ribonucleotide synthetase
|
| 46 | c2recB_ |
|
not modelled |
73.5 |
13 |
PDB header:helicase
PDB COMPND:
|
| 47 | c3brqA_ |
|
not modelled |
73.1 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator ascg;
PDBTitle: crystal structure of the escherichia coli transcriptional repressor2 ascg
|
| 48 | c3n7uD_ |
|
not modelled |
72.3 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:formate dehydrogenase;
PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
|
| 49 | c3qk7C_ |
|
not modelled |
71.1 |
15 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcriptional regulator from yersinia2 pestis biovar microtus str. 91001
|
| 50 | c2yvqA_ |
|
not modelled |
70.7 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 51 | d1miob_ |
|
not modelled |
70.2 |
13 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 52 | c1lkzB_ |
|
not modelled |
70.0 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase a;
PDBTitle: crystal structure of d-ribose-5-phosphate isomerase (rpia)2 from escherichia coli.
|
| 53 | d1t0ia_ |
|
not modelled |
69.8 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 54 | c3d8uA_ |
|
not modelled |
69.3 |
10 |
PDB header:transcription regulator
Chain: A: PDB Molecule:purr transcriptional regulator;
PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
|
| 55 | c1kjjA_ |
|
not modelled |
69.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase 2;
PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
|
| 56 | c2zroA_ |
|
not modelled |
68.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein reca;
PDBTitle: msreca adp form iv
|
| 57 | c2eghA_ |
|
not modelled |
68.8 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
|
| 58 | c3npgD_ |
|
not modelled |
68.6 |
14 |
PDB header:unknown function
Chain: D: PDB Molecule:uncharacterized duf364 family protein;
PDBTitle: crystal structure of a protein with unknown function from duf3642 family (ph1506) from pyrococcus horikoshii at 2.70 a resolution
|
| 59 | d2a5la1 |
|
not modelled |
68.1 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 60 | c3bazA_ |
|
not modelled |
67.7 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyphenylpyruvate reductase;
PDBTitle: structure of hydroxyphenylpyruvate reductase from coleus blumei in2 complex with nadp+
|
| 61 | c3ezxA_ |
|
not modelled |
67.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
| 62 | c3k4hA_ |
|
not modelled |
67.1 |
8 |
PDB header:transcription regulator
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator laci from2 bacillus cereus subsp. cytotoxis nvh 391-98
|
| 63 | c3gebC_ |
|
not modelled |
66.5 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:eyes absent homolog 2;
PDBTitle: crystal structure of edeya2
|
| 64 | c3g1wB_ |
|
not modelled |
65.9 |
7 |
PDB header:transport protein
Chain: B: PDB Molecule:sugar abc transporter;
PDBTitle: crystal structure of sugar abc transporter (sugar-binding protein)2 from bacillus halodurans
|
| 65 | c3k9cA_ |
|
not modelled |
65.9 |
12 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family protein;
PDBTitle: crystal structure of laci transcriptional regulator from rhodococcus2 species.
|
| 66 | c2issF_ |
|
not modelled |
65.4 |
32 |
PDB header:lyase, transferase
Chain: F: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: structure of the plp synthase holoenzyme from thermotoga maritima
|
| 67 | d1o8bb1 |
|
not modelled |
65.2 |
13 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 68 | d2nv0a1 |
|
not modelled |
64.8 |
20 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 69 | d1wl8a1 |
|
not modelled |
64.3 |
11 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 70 | c3gt7A_ |
|
not modelled |
62.9 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of signal receiver domain of signal2 transduction histidine kinase from syntrophus3 aciditrophicus
|
| 71 | d1qo0a_ |
|
not modelled |
62.7 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 72 | c2zayA_ |
|
not modelled |
62.6 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator from desulfuromonas2 acetoxidans
|
| 73 | c3uifA_ |
|
not modelled |
62.5 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:sulfonate abc transporter, periplasmic sulfonate-binding
PDBTitle: crystal structure of putative sulfonate abc transporter, periplasmic2 sulfonate-binding protein ssua from methylobacillus flagellatus kt
|
| 74 | c1m0sA_ |
|
not modelled |
62.0 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: northeast structural genomics consortium (nesg id ir21)
|
| 75 | d1fmfa_ |
|
not modelled |
61.5 |
9 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 76 | d1zesa1 |
|
not modelled |
61.5 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 77 | c1bvyF_ |
|
not modelled |
61.3 |
13 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:protein (cytochrome p450 bm-3);
PDBTitle: complex of the heme and fmn-binding domains of the2 cytochrome p450(bm-3)
|
| 78 | d1bvyf_ |
|
not modelled |
61.3 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 79 | c2dwcB_ |
|
not modelled |
61.2 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:433aa long hypothetical phosphoribosylglycinamide formyl
PDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
|
| 80 | c3fwzA_ |
|
not modelled |
60.9 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ybal;
PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
|
| 81 | c3hheA_ |
|
not modelled |
60.9 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from bartonella2 henselae
|
| 82 | c3clkB_ |
|
not modelled |
60.8 |
8 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcription regulator;
PDBTitle: crystal structure of a transcription regulator from lactobacillus2 plantarum
|
| 83 | d1vmea1 |
|
not modelled |
60.6 |
14 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 84 | d1p90a_ |
|
not modelled |
60.3 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:Nitrogenase accessory factor |
| 85 | c3e4rA_ |
|
not modelled |
59.9 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein;
PDBTitle: crystal structure of the alkanesulfonate binding protein2 (ssua) from the phytopathogenic bacteria xanthomonas3 axonopodis pv. citri bound to hepes
|
| 86 | d2hmva1 |
|
not modelled |
59.5 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 87 | d1mioa_ |
|
not modelled |
59.4 |
12 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 88 | d1d4aa_ |
|
not modelled |
59.3 |
30 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 89 | c3brsA_ |
|
not modelled |
58.5 |
5 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of sugar transporter from clostridium2 phytofermentans
|
| 90 | c1uj6A_ |
|
not modelled |
57.9 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of thermus thermophilus ribose-5-phosphate isomerase2 complexed with arabinose-5-phosphate
|
| 91 | c3gv0A_ |
|
not modelled |
57.8 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
|
| 92 | c3l6uA_ |
|
not modelled |
57.8 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system periplasmic
PDBTitle: crystal structure of abc-type sugar transport system,2 periplasmic component from exiguobacterium sibiricum
|
| 93 | c3a0rB_ |
|
not modelled |
57.7 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
|
| 94 | d1qrda_ |
|
not modelled |
57.1 |
24 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 95 | c1xtzA_ |
|
not modelled |
56.7 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
|
| 96 | d1xi8a3 |
|
not modelled |
56.0 |
11 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 97 | c3l7oB_ |
|
not modelled |
55.7 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
| 98 | c2zkiH_ |
|
not modelled |
55.1 |
15 |
PDB header:transcription
Chain: H: PDB Molecule:199aa long hypothetical trp repressor binding
PDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
|
| 99 | c3u7jA_ |
|
not modelled |
54.9 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
|
| 100 | c2rjoA_ |
|
not modelled |
54.5 |
8 |
PDB header:signaling protein
Chain: A: PDB Molecule:twin-arginine translocation pathway signal protein;
PDBTitle: crystal structure of twin-arginine translocation pathway signal2 protein from burkholderia phytofirmans
|
| 101 | c2xdqA_ |
|
not modelled |
54.2 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-2 chlb)2 complex
|
| 102 | c2q5cA_ |
|
not modelled |
54.2 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 103 | d2a9va1 |
|
not modelled |
53.3 |
26 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 104 | d1a9xa2 |
|
not modelled |
52.6 |
18 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 105 | c3h5oB_ |
|
not modelled |
52.5 |
11 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator gntr;
PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
|
| 106 | c3d3jA_ |
|
not modelled |
52.4 |
9 |
PDB header:protein binding
Chain: A: PDB Molecule:enhancer of mrna-decapping protein 3;
PDBTitle: crystal structure of human edc3p
|
| 107 | c2ioyB_ |
|
not modelled |
52.4 |
12 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:periplasmic sugar-binding protein;
PDBTitle: crystal structure of thermoanaerobacter tengcongensis2 ribose binding protein
|
| 108 | c1xp8A_ |
|
not modelled |
52.0 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:reca protein;
PDBTitle: "deinococcus radiodurans reca in complex with atp-gamma-s"
|
| 109 | c3l4bG_ |
|
not modelled |
50.3 |
8 |
PDB header:transport protein
Chain: G: PDB Molecule:trka k+ channel protien tm1088b;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
|
| 110 | c1vkzA_ |
|
not modelled |
49.6 |
7 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of phosphoribosylamine--glycine ligase (tm1250) from2 thermotoga maritima at 2.30 a resolution
|
| 111 | c1e1cA_ |
|
not modelled |
49.6 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
| 112 | c3kwmC_ |
|
not modelled |
49.3 |
16 |
PDB header:isomerase
Chain: C: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-isomerase a
|
| 113 | d1dxya2 |
|
not modelled |
48.7 |
10 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 114 | c3icpA_ |
|
not modelled |
48.4 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of udp-galactose 4-epimerase
|
| 115 | c3f2vA_ |
|
not modelled |
48.3 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:general stress protein 14;
PDBTitle: crystal structure of the general stress protein 142 (tde0354) in complex with fmn from treponema denticola,3 northeast structural genomics consortium target tdr58.
|
| 116 | d1vkza2 |
|
not modelled |
48.0 |
7 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 117 | c2rgyA_ |
|
not modelled |
47.9 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of transcriptional regulator of laci family from2 burkhoderia phymatum
|
| 118 | d1kjqa2 |
|
not modelled |
47.8 |
11 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 119 | c2ywjA_ |
|
not modelled |
47.4 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: crystal structure of uncharacterized conserved protein from2 methanocaldococcus jannaschii
|
| 120 | c2p5uC_ |
|
not modelled |
47.4 |
24 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
|