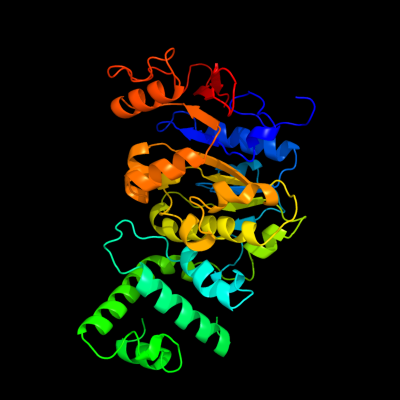
1 d1e9ra_
100.0
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)2 c2iusB_
99.7
22
PDB header: membrane proteinChain: B: PDB Molecule: dna translocase ftsk;PDBTitle: e. coli ftsk motor domain
3 c2iutA_
99.7
21
PDB header: membrane proteinChain: A: PDB Molecule: dna translocase ftsk;PDBTitle: p. aeruginosa ftsk motor domain, dimeric
4 c2iuuE_
99.6
21
PDB header: membrane proteinChain: E: PDB Molecule: dna translocase ftsk;PDBTitle: p. aeruginosa ftsk motor domain, hexamer
5 d1ji0a_
98.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like6 d1vpla_
98.0
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like7 c2nq2C_
98.0
16
PDB header: metal transportChain: C: PDB Molecule: hypothetical abc transporter atp-binding proteinPDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
8 c2yz2B_
97.9
15
PDB header: hydrolaseChain: B: PDB Molecule: putative abc transporter atp-binding protein tm_0222;PDBTitle: crystal structure of the abc transporter in the cobalt transport2 system
9 c1oxtB_
97.9
14
PDB header: transport proteinChain: B: PDB Molecule: abc transporter, atp binding protein;PDBTitle: crystal structure of glcv, the abc-atpase of the glucose abc2 transporter from sulfolobus solfataricus
10 c2olkD_
97.9
13
PDB header: hydrolaseChain: D: PDB Molecule: amino acid abc transporter;PDBTitle: abc protein artp in complex with adp-beta-s
11 c3fvqB_
97.8
16
PDB header: hydrolaseChain: B: PDB Molecule: fe(3+) ions import atp-binding protein fbpc;PDBTitle: crystal structure of the nucleotide binding domain fbpc2 complexed with atp
12 d1g6ha_
97.8
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like13 c1z47B_
97.8
16
PDB header: ligand binding proteinChain: B: PDB Molecule: putative abc-transporter atp-binding protein;PDBTitle: structure of the atpase subunit cysa of the putative2 sulfate atp-binding cassette (abc) transporter from3 alicyclobacillus acidocaldarius
14 c2r2aB_
97.8
13
PDB header: toxinChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of n-terminal domain of zonular occludens toxin from2 neisseria meningitidis
15 d1b0ua_
97.8
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like16 d2qy9a2
97.7
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like17 d1vmaa2
97.7
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like18 c2xgjA_
97.7
23
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: structure of mtr4, a dexh helicase involved in nuclear rna2 processing and surveillance
19 c3l9oA_
97.6
22
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: crystal structure of mtr4, a co-factor of the nuclear exosome
20 d1np6a_
97.6
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like21 d1okkd2
not modelled
97.6
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like22 c2w0mA_
not modelled
97.6
20
PDB header: unknown functionChain: A: PDB Molecule: sso2452;PDBTitle: crystal structure of sso2452 from sulfolobus solfataricus2 p2
23 d1g6oa_
not modelled
97.6
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)24 d2i3ba1
not modelled
97.6
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)25 c2yl4A_
not modelled
97.6
17
PDB header: membrane proteinChain: A: PDB Molecule: atp-binding cassette sub-family b member 10,PDBTitle: structure of the human mitochondrial abc transporter, abcb10
26 d1l7vc_
not modelled
97.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like27 d1p9ra_
not modelled
97.5
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)28 d3dhwc1
not modelled
97.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like29 c4a4zA_
not modelled
97.5
16
PDB header: hydrolaseChain: A: PDB Molecule: antiviral helicase ski2;PDBTitle: crystal structure of the s. cerevisiae dexh helicase ski2 bound to2 amppnp
30 c2f1rA_
not modelled
97.5
26
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin-guanine dinucleotide biosynthesisPDBTitle: crystal structure of molybdopterin-guanine biosynthesis2 protein b (mobb)
31 d1qzxa3
not modelled
97.5
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like32 c2gzaB_
not modelled
97.5
20
PDB header: hydrolaseChain: B: PDB Molecule: type iv secretion system protein virb11;PDBTitle: crystal structure of the virb11 atpase from the brucella suis type iv2 secretion system in complex with sulphate
33 c3jvvA_
not modelled
97.4
12
PDB header: atp binding proteinChain: A: PDB Molecule: twitching mobility protein;PDBTitle: crystal structure of p. aeruginosa pilt with bound amp-pcp
34 d1ls1a2
not modelled
97.4
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like35 c2oaq1_
not modelled
97.4
17
PDB header: hydrolaseChain: 1: PDB Molecule: type ii secretion system protein;PDBTitle: crystal structure of the archaeal secretion atpase gspe in complex2 with phosphate
36 c3a4mB_
not modelled
97.4
18
PDB header: transferaseChain: B: PDB Molecule: l-seryl-trna(sec) kinase;PDBTitle: crystal structure of archaeal o-phosphoseryl-trna(sec)2 kinase
37 c2cnwF_
not modelled
97.3
21
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
38 c2ja1A_
not modelled
97.3
21
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: thymidine kinase from b. cereus with ttp bound as phosphate2 donor.
39 c2j7pA_
not modelled
97.3
16
PDB header: signal recognitionChain: A: PDB Molecule: signal recognition particle protein;PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
40 c2qy9A_
not modelled
97.3
20
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
41 d1rz3a_
not modelled
97.3
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase42 c1zu4A_
not modelled
97.2
17
PDB header: protein transportChain: A: PDB Molecule: ftsy;PDBTitle: crystal structure of ftsy from mycoplasma mycoides- space2 group p21212
43 c2gszE_
not modelled
97.2
19
PDB header: protein transportChain: E: PDB Molecule: twitching motility protein pilt;PDBTitle: structure of a. aeolicus pilt with 6 monomers per2 asymmetric unit
44 c1vmaA_
not modelled
97.2
24
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
45 c3b9qA_
not modelled
97.1
16
PDB header: protein transportChain: A: PDB Molecule: chloroplast srp receptor homolog, alpha subunitPDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
46 c2og2A_
not modelled
97.1
17
PDB header: protein transportChain: A: PDB Molecule: putative signal recognition particle receptor;PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
47 d2qm8a1
not modelled
97.1
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like48 c2yhsA_
not modelled
97.1
21
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the e. coli srp receptor ftsy
49 d1byia_
not modelled
97.1
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like50 d1tf7a2
not modelled
97.0
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)51 d1tf7a1
not modelled
97.0
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)52 c2dr3A_
not modelled
97.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0273 protein ph0284;PDBTitle: crystal structure of reca superfamily atpase ph0284 from pyrococcus2 horikoshii ot3
53 c3c8uA_
not modelled
97.0
31
PDB header: transferaseChain: A: PDB Molecule: fructokinase;PDBTitle: crystal structure of putative fructose transport system kinase2 (yp_612366.1) from silicibacter sp. tm1040 at 1.95 a resolution
54 c3dmdA_
not modelled
97.0
26
PDB header: transport proteinChain: A: PDB Molecule: signal recognition particle receptor;PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
55 c3hr8A_
not modelled
97.0
26
PDB header: recombinationChain: A: PDB Molecule: protein reca;PDBTitle: crystal structure of thermotoga maritima reca
56 d1nksa_
not modelled
97.0
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases57 c2recB_
not modelled
96.9
21
PDB header: helicasePDB COMPND: 58 c1xp8A_
not modelled
96.9
21
PDB header: dna binding proteinChain: A: PDB Molecule: reca protein;PDBTitle: "deinococcus radiodurans reca in complex with atp-gamma-s"
59 c1xx6B_
not modelled
96.9
18
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: x-ray structure of clostridium acetobutylicum thymidine kinase with2 adp. northeast structural genomics target car26.
60 c1u9iA_
not modelled
96.9
22
PDB header: circadian clock proteinChain: A: PDB Molecule: kaic;PDBTitle: crystal structure of circadian clock protein kaic with phosphorylation2 sites
61 c3nxsA_
not modelled
96.9
24
PDB header: transport proteinChain: A: PDB Molecule: lao/ao transport system atpase;PDBTitle: crystal structure of lao/ao transport system from mycobacterium2 smegmatis bound to gdp
62 c2wwwB_
not modelled
96.9
20
PDB header: transport proteinChain: B: PDB Molecule: methylmalonic aciduria type a protein,PDBTitle: crystal structure of methylmalonic acidemia type a protein
63 c3tqcB_
not modelled
96.9
14
PDB header: transferaseChain: B: PDB Molecule: pantothenate kinase;PDBTitle: structure of the pantothenate kinase (coaa) from coxiella burnetii
64 c3cmvG_
not modelled
96.9
21
PDB header: recombinationChain: G: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
65 d1u94a1
not modelled
96.9
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)66 d1rkba_
not modelled
96.9
34
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases67 d1mo6a1
not modelled
96.9
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)68 c2wwiC_
not modelled
96.9
10
PDB header: transferaseChain: C: PDB Molecule: thymidilate kinase, putative;PDBTitle: plasmodium falciparum thymidylate kinase in complex with2 aztmp and adp
69 c2eyuA_
not modelled
96.9
21
PDB header: protein transportChain: A: PDB Molecule: twitching motility protein pilt;PDBTitle: the crystal structure of the c-terminal domain of aquifex2 aeolicus pilt
70 d1ye8a1
not modelled
96.9
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)71 c3b85A_
not modelled
96.8
24
PDB header: hydrolaseChain: A: PDB Molecule: phosphate starvation-inducible protein;PDBTitle: crystal structure of predicted phosphate starvation-induced atpase2 phoh2 from corynebacterium glutamicum
72 d1l8qa2
not modelled
96.8
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain73 c2zroA_
not modelled
96.8
21
PDB header: hydrolaseChain: A: PDB Molecule: protein reca;PDBTitle: msreca adp form iv
74 d1ki9a_
not modelled
96.8
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases75 c2gesA_
not modelled
96.8
14
PDB header: transferaseChain: A: PDB Molecule: pantothenate kinase;PDBTitle: pantothenate kinase from mycobacterium tuberculosis (mtpank) in2 complex with a coenzyme a derivative, form-i (rt)
76 c2z0hA_
not modelled
96.8
24
PDB header: transferaseChain: A: PDB Molecule: thymidylate kinase;PDBTitle: crystal structure of thymidylate kinase in complex with dtdp2 and adp from thermotoga maritima
77 d1sq5a_
not modelled
96.8
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase78 d1xp8a1
not modelled
96.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)79 c3oiyB_
not modelled
96.8
13
PDB header: isomeraseChain: B: PDB Molecule: reverse gyrase helicase domain;PDBTitle: helicase domain of reverse gyrase from thermotoga maritima
80 c2iy3A_
not modelled
96.8
16
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein ffh;PDBTitle: structure of the e. coli signal recognition particle2 bound to a translating ribosome
81 d1ubea1
not modelled
96.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)82 c3asyB_
not modelled
96.8
33
PDB header: transferaseChain: B: PDB Molecule: uridine kinase;PDBTitle: ligand-free structure of uridine kinase from thermus thermophilus hb8
83 c3of5A_
not modelled
96.8
18
PDB header: ligaseChain: A: PDB Molecule: dethiobiotin synthetase;PDBTitle: crystal structure of a dethiobiotin synthetase from francisella2 tularensis subsp. tularensis schu s4
84 d1xjca_
not modelled
96.8
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like85 d1khta_
not modelled
96.8
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases86 c2gksB_
not modelled
96.7
21
PDB header: transferaseChain: B: PDB Molecule: bifunctional sat/aps kinase;PDBTitle: crystal structure of the bi-functional atp sulfurylase-aps kinase from2 aquifex aeolicus, a chemolithotrophic thermophile
87 d1odfa_
not modelled
96.7
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase88 d1yrba1
not modelled
96.7
35
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like89 c3p1jC_
not modelled
96.7
27
PDB header: hydrolaseChain: C: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of human gtpase imap family member 2 in the2 nucleotide-free state
90 c3lfuA_
not modelled
96.7
19
PDB header: hydrolaseChain: A: PDB Molecule: dna helicase ii;PDBTitle: crystal structure of e. coli uvrd
91 c3bosA_
not modelled
96.7
13
PDB header: hydrolase regulator,dna binding proteinChain: A: PDB Molecule: putative dna replication factor;PDBTitle: crystal structure of a putative dna replication regulator hda2 (sama_1916) from shewanella amazonensis sb2b at 1.75 a resolution
92 d1bifa1
not modelled
96.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, kinase domain93 c2hcbC_
not modelled
96.7
18
PDB header: replicationChain: C: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: structure of amppcp-bound dnaa from aquifex aeolicus
94 c3dm5A_
not modelled
96.7
26
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
95 c1zuiA_
not modelled
96.7
30
PDB header: transferaseChain: A: PDB Molecule: shikimate kinase;PDBTitle: structural basis for shikimate-binding specificity of helicobacter2 pylori shikimate kinase
96 c2pbrB_
not modelled
96.7
27
PDB header: transferaseChain: B: PDB Molecule: thymidylate kinase;PDBTitle: crystal structure of thymidylate kinase (aq_969) from aquifex aeolicus2 vf5
97 c3lv8A_
not modelled
96.6
25
PDB header: transferaseChain: A: PDB Molecule: thymidylate kinase;PDBTitle: 1.8 angstrom resolution crystal structure of a thymidylate kinase2 (tmk) from vibrio cholerae o1 biovar eltor str. n16961 in complex3 with tmp, thymidine-5'-diphosphate and adp
98 d1x6va3
not modelled
96.6
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Adenosine-5'phosphosulfate kinase (APS kinase)99 c2grjH_
not modelled
96.6
30
PDB header: transferaseChain: H: PDB Molecule: dephospho-coa kinase;PDBTitle: crystal structure of dephospho-coa kinase (ec 2.7.1.24)2 (dephosphocoenzyme a kinase) (tm1387) from thermotoga maritima at3 2.60 a resolution
100 c2qagB_
not modelled
96.6
32
PDB header: cell cycle, structural proteinChain: B: PDB Molecule: septin-6;PDBTitle: crystal structure of human septin trimer 2/6/7
101 c2npiB_
not modelled
96.6
23
PDB header: transcriptionChain: B: PDB Molecule: protein clp1;PDBTitle: clp1-atp-pcf11 complex
102 d1gkub1
not modelled
96.6
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Helicase-like "domain" of reverse gyrase103 c2qq0B_
not modelled
96.6
16
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: thymidine kinase from thermotoga maritima in complex with2 thymidine + appnhp
104 d1fnna2
not modelled
96.6
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain105 d2p67a1
not modelled
96.6
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like106 c2qa5A_
not modelled
96.6
41
PDB header: cell cycle, structural proteinChain: A: PDB Molecule: septin-2;PDBTitle: crystal structure of sept2 g-domain
107 d1k6ma1
not modelled
96.5
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, kinase domain108 c3t5dC_
not modelled
96.5
41
PDB header: signaling proteinChain: C: PDB Molecule: septin-7;PDBTitle: crystal structure of septin 7 in complex with gdp
109 c2kjqA_
not modelled
96.5
30
PDB header: replicationChain: A: PDB Molecule: dnaa-related protein;PDBTitle: solution structure of protein nmb1076 from neisseria meningitidis.2 northeast structural genomics consortium target mr101b.
110 c3bh0A_
not modelled
96.5
13
PDB header: replicationChain: A: PDB Molecule: dnab-like replicative helicase;PDBTitle: atpase domain of g40p
111 d1ny5a2
not modelled
96.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain112 c3md0A_
not modelled
96.5
25
PDB header: transport proteinChain: A: PDB Molecule: arginine/ornithine transport system atpase;PDBTitle: crystal structure of arginine/ornithine transport system2 atpase from mycobacterium tuberculosis bound to gdp (a ras-3 like gtpase superfamily protein)
113 c3be4A_
not modelled
96.5
21
PDB header: transferaseChain: A: PDB Molecule: adenylate kinase;PDBTitle: crystal structure of cryptosporidium parvum adenylate kinase cgd5_3360
114 d1cr2a_
not modelled
96.5
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)115 d2afhe1
not modelled
96.5
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like116 c2plrB_
not modelled
96.5
20
PDB header: transferaseChain: B: PDB Molecule: probable thymidylate kinase;PDBTitle: crystal structure of dtmp kinase (st1543) from sulfolobus tokodaii2 strain7
117 c3lxwA_
not modelled
96.5
22
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 1;PDBTitle: crystal structure of human gtpase imap family member 1
118 d1e4va1
not modelled
96.5
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases119 d1akya1
not modelled
96.5
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases120 d1zina1
not modelled
96.5
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases