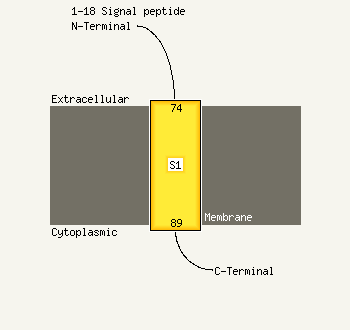
1 d1roaa_
100.0
56
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins
2 d1r4ca_
100.0
97
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins
3 c2ch9A_
100.0
33
PDB header: inhibitorChain: A: PDB Molecule: cystatin f;PDBTitle: crystal structure of dimeric human cystatin f
4 d1cewi_
100.0
44
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins
5 c4it7C_
100.0
27
PDB header: hydrolase inhibitorChain: C: PDB Molecule: cpi;PDBTitle: crystal structure of al-cpi
6 c3mwzA_
100.0
25
PDB header: hydrolase inhibitorChain: A: PDB Molecule: sialostatin l2;PDBTitle: crystal structure of the selenomethionine derivative of the l 22,47,2 100 m mutant of sialostatin l2
7 c3l0rA_
100.0
23
PDB header: hydrolase inhibitorChain: A: PDB Molecule: cystatin-2;PDBTitle: crystal structure of salivary cystatin from the soft tick ornithodoros2 moubata
8 c4lziA_
99.9
24
PDB header: hydrolase inhibitorChain: A: PDB Molecule: multicystatin;PDBTitle: characterization of solanum tuberosum multicystatin and significance2 of core domains
9 c2l4vA_
99.9
30
PDB header: hydrolase inhibitorChain: A: PDB Molecule: cystatin;PDBTitle: three dimensional structure of pineapple cystatin
10 c3imaD_
99.8
30
PDB header: hydrolase/hydrolase inhibitorChain: D: PDB Molecule: cysteine proteinase inhibitor;PDBTitle: complex strcuture of tarocystatin and papain
11 d1eqka_
99.8
23
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins
12 c2w9pC_
99.8
20
PDB header: hydrolase inhibitorChain: C: PDB Molecule: multicystatin;PDBTitle: crystal structure of potato multicystatin
13 c4eycA_
99.8
19
PDB header: unknown functionChain: A: PDB Molecule: cathelicidin antimicrobial peptide;PDBTitle: crystal structure of the cathelin-like domain of human cathelicidin2 ll-37 (hcld)
14 c3ul5C_
99.7
25
PDB header: hydrolase inhibitorChain: C: PDB Molecule: canecystatin-1;PDBTitle: saccharum officinarum canecystatin-1 in space group c2221
15 d2b4cg1
99.6
80
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core
16 d1stfi_
99.5
24
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins
17 c3j5mI_
99.5
72
PDB header: viral protein/immune systemChain: I: PDB Molecule: bg505 sosip gp120;PDBTitle: cryo-em structure of the bg505 sosip.664 hiv-1 env trimer with 3 pgv042 fabs
18 c1ce4A_
99.0
85
PDB header: viral proteinChain: A: PDB Molecule: protein (v3 loop of hiv-1 envelope protein);PDBTitle: conformational model for the consensus v3 loop of the2 envelope protein gp120 of hiv-1
19 c3ngbI_
98.7
62
PDB header: viral protein/immune systemChain: I: PDB Molecule: envelope glycoprotein gp160;PDBTitle: crystal structure of broadly and potently neutralizing antibody vrc012 in complex with hiv-1 gp120
20 d1nb5i_
98.6
24
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins
21 d1kwia_
not modelled
97.8
22
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cathelicidin motif
22 c4i3rG_
not modelled
97.7
51
PDB header: viral protein/immune systemChain: G: PDB Molecule: outer domain of hiv-1 gp120 (ker2018 od4.2.2);PDBTitle: crystal structure of the outer domain of hiv-1 gp120 in complex with2 vrc-pg04 space group p3221
23 d2bo9b1
not modelled
97.3
17
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Latexin-like
24 c2bo9B_
not modelled
96.9
18
PDB header: hydrolaseChain: B: PDB Molecule: human latexin;PDBTitle: human carboxypeptidase a4 in complex with human latexin.
25 c3rjqA_
not modelled
96.8
84
PDB header: viral protein/immune systemChain: A: PDB Molecule: c186 gp120;PDBTitle: crystal structure of anti-hiv llama vhh antibody a12 in complex with2 c186 gp120
26 c3jwdA_
not modelled
96.5
91
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 gp120 envelope glycoprotein;PDBTitle: structure of hiv-1 gp120 with gp41-interactive region: layered2 architecture and basis of conformational mobility
27 c3dnlB_
not modelled
96.3
82
PDB header: viral proteinChain: B: PDB Molecule: hiv-1 envelope glycoprotein gp120;PDBTitle: molecular structure for the hiv-1 gp120 trimer in the b12-2 bound state
28 d2nxya1
not modelled
96.3
79
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core
29 d1yymg1
not modelled
95.9
64
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core
30 c4ccvA_
not modelled
93.6
21
PDB header: blood clottingChain: A: PDB Molecule: histidine-rich glycoprotein;PDBTitle: crystal structure of histidine-rich glycoprotein n2 domain2 reveals redox activity at an interdomain disulfide bridge:3 implications for the regulation of angiogenesis
31 c3mlwQ_
not modelled
92.1
100
PDB header: immune systemChain: Q: PDB Molecule: hiv-1 gp120 third variable region (v3) crown;PDBTitle: crystal structure of anti-hiv-1 v3 fab 1006-15d in complex with an mn2 v3 peptide
32 c3ujiP_
not modelled
92.0
100
PDB header: immune systemChain: P: PDB Molecule: envelope glycoprotein gp160;PDBTitle: crystal structure of anti-hiv-1 v3 fab 2558 in complex with mn peptide
33 c3ujjP_
not modelled
91.8
65
PDB header: immune systemChain: P: PDB Molecule: gp120;PDBTitle: crystal structure of anti-hiv-1 v3 fab 4025 in complex with con a2 peptide
34 c2b1aP_
not modelled
89.2
75
PDB header: immune systemChain: P: PDB Molecule: ug1033 peptide of exterior membrane glycoprotein gp120;PDBTitle: crystal structure analysis of anti-hiv-1 v3 fab 2219 in complex with2 ug1033 peptide
35 c3mlwP_
not modelled
88.5
100
PDB header: immune systemChain: P: PDB Molecule: hiv-1 gp120 third variable region (v3) crown;PDBTitle: crystal structure of anti-hiv-1 v3 fab 1006-15d in complex with an mn2 v3 peptide
36 c2kxgA_
not modelled
80.5
21
PDB header: hydrolase inhibitorChain: A: PDB Molecule: aspartic protease inhibitor;PDBTitle: the solution structure of the squash aspartic acid proteinase2 inhibitor (sqapi)
37 c2bf1A_
not modelled
80.0
4
PDB header: virus proteinChain: A: PDB Molecule: exterior membrane glycoprotein gp120;PDBTitle: structure of an unliganded and fully-glycosylated siv gp1202 envelope glycoprotein
38 c1y4cA_
73.1
12
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
39 c2zv4O_
not modelled
58.4
17
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
40 c1acyP_
not modelled
43.8
91
PDB header: complex(antibody/hiv-1 fragment)Chain: P: PDB Molecule: hiv-1 gp120 (mn isolate);PDBTitle: crystal structure of the principal neutralizing site of hiv-2 1
41 c3mlrP_
not modelled
40.0
50
PDB header: immune systemChain: P: PDB Molecule: hiv-1 gp120 third variable region (v3) crown;PDBTitle: crystal structure of anti-hiv-1 v3 fab 2557 in complex with a ny5 v32 peptide
42 c2w82C_
not modelled
31.4
24
PDB header: replication inhibitorChain: C: PDB Molecule: orf18;PDBTitle: the structure of arda
43 d1fcda3
not modelled
29.7
25
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
44 c2e4wA_
not modelled
18.0
26
PDB header: signaling proteinChain: A: PDB Molecule: metabotropic glutamate receptor 3;PDBTitle: crystal structure of the extracellular region of the group ii2 metabotropic glutamate receptor complexed with 1s,3s-acpd
45 c3fewX_
not modelled
17.4
21
PDB header: immune systemChain: X: PDB Molecule: colicin s4;PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
46 c3mlvQ_
not modelled
15.5
53
PDB header: immune systemChain: Q: PDB Molecule: hiv-1 gp120 third variable region (v3) crown;PDBTitle: crystal structure of anti-hiv-1 v3 fab 2557 in complex with an nof v32 peptide
47 c3mlvP_
not modelled
15.5
53
PDB header: immune systemChain: P: PDB Molecule: hiv-1 gp120 third variable region (v3) crown;PDBTitle: crystal structure of anti-hiv-1 v3 fab 2557 in complex with an nof v32 peptide
48 c3oqtP_
not modelled
14.7
21
PDB header: flavoproteinChain: P: PDB Molecule: rv1498a protein;PDBTitle: crystal structure of rv1498a protein from mycobacterium tuberculosis
49 c3p3wC_
not modelled
13.8
23
PDB header: transport proteinChain: C: PDB Molecule: glutamate receptor 3;PDBTitle: structure of a dimeric glua3 n-terminal domain (ntd) at 4.2 a2 resolution
50 c3monF_
not modelled
13.6
23
PDB header: sweet-tasting proteinChain: F: PDB Molecule: monellin;PDBTitle: crystal structures of two intensely sweet proteins
51 d1wkaa1
not modelled
12.5
30
Fold: ValRS/IleRS/LeuRS editing domainSuperfamily: ValRS/IleRS/LeuRS editing domainFamily: ValRS/IleRS/LeuRS editing domain
52 c2vxaL_
not modelled
11.8
29
PDB header: flavoproteinChain: L: PDB Molecule: dodecin;PDBTitle: h.halophila dodecin in complex with riboflavin
53 c3onrI_
not modelled
11.3
17
PDB header: metal binding proteinChain: I: PDB Molecule: protein transport protein sece2;PDBTitle: crystal structure of the calcium chelating immunodominant antigen,2 calcium dodecin (rv0379),from mycobacterium tuberculosis with a novel3 calcium-binding site
54 c2hlqA_
not modelled
11.0
24
PDB header: transferaseChain: A: PDB Molecule: bone morphogenetic protein receptor type-2;PDBTitle: crystal structure of the extracellular domain of the type2 ii bmp receptor
55 c3j20F_
not modelled
10.6
10
PDB header: ribosomeChain: F: PDB Molecule: 30s ribosomal protein s5p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
56 d2ux9a1
not modelled
10.5
27
Fold: Dodecin subunit-likeSuperfamily: Dodecin-likeFamily: Dodecin-like
57 c4uqqD_
not modelled
9.1
17
PDB header: transport proteinChain: D: PDB Molecule: glutamate receptor ionotropic, kainate 2;PDBTitle: electron density map of gluk2 desensitized state in2 complex with 2s,4r-4-methylglutamate
58 c4f11A_
not modelled
7.9
20
PDB header: signaling proteinChain: A: PDB Molecule: gamma-aminobutyric acid type b receptor subunit 2;PDBTitle: crystal structure of the extracellular domain of human gaba(b)2 receptor gbr2
59 d2o9ux1
not modelled
7.6
23
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Monellin
60 c3sajB_
not modelled
7.6
16
PDB header: transport proteinChain: B: PDB Molecule: glutamate receptor 1;PDBTitle: crystal structure of glutamate receptor glua1 amino terminal domain
61 d1ntga_
not modelled
7.4
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Myf domain
62 c3heiF_
not modelled
7.3
27
PDB header: transferase/signaling proteinChain: F: PDB Molecule: ephrin-a1;PDBTitle: ligand recognition by a-class eph receptors: crystal structures of the2 epha2 ligand-binding domain and the epha2/ephrin-a1 complex
63 c1rrqA_
not modelled
6.9
36
PDB header: hydrolase/dnaChain: A: PDB Molecule: muty;PDBTitle: muty adenine glycosylase in complex with dna containing an2 a:oxog pair
64 c1zzaA_
not modelled
6.8
24
PDB header: membrane proteinChain: A: PDB Molecule: stannin;PDBTitle: solution nmr structure of the membrane protein stannin
65 d1t1ra3
not modelled
6.7
57
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
66 d2nqda1
not modelled
6.6
50
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ICP-likeFamily: ICP-like
67 d2p3ra1
not modelled
6.5
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glycerol kinase
68 c2qzvB_
not modelled
6.4
11
PDB header: structural proteinChain: B: PDB Molecule: major vault protein;PDBTitle: draft crystal structure of the vault shell at 9 angstroms2 resolution
69 d1mn4a_
not modelled
6.4
25
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: DNA-binding domain from NDT80
70 d1vdda_
not modelled
6.3
14
Fold: Recombination protein RecRSuperfamily: Recombination protein RecRFamily: Recombination protein RecR
71 d1iyka1
not modelled
6.3
9
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT
72 c3om1A_
not modelled
6.3
21
PDB header: membrane proteinChain: A: PDB Molecule: glutamate receptor gluk5 (ka2);PDBTitle: crystal structure of the gluk5 (ka2) atd dimer at 1.7 angstrom2 resolution
73 c4kzzC_
not modelled
6.2
20
PDB header: ribosomeChain: C: PDB Molecule: 40s ribosomal protein s2;PDBTitle: rabbit 40s ribosomal subunit in complex with mrna, initiator trna and2 eif1a
74 d1dx5i1
not modelled
6.2
29
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: EGF-type module
75 c3qwmA_
not modelled
6.1
9
PDB header: signaling proteinChain: A: PDB Molecule: iq motif and sec7 domain-containing protein 1;PDBTitle: crystal structure of gep100, the plextrin homology domain of iq motif2 and sec7 domain-containing protein 1 isoform a
76 d1wlta1
not modelled
6.1
17
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
77 c2kkmA_
not modelled
6.0
16
PDB header: translationChain: A: PDB Molecule: translation machinery-associated protein 16;PDBTitle: solution nmr structure of yeast protein yor252w [residues2 38-178]: northeast structural genomics consortium target3 yt654
78 d1m5ha2
not modelled
5.9
23
Fold: Ferredoxin-likeSuperfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferaseFamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase
79 c3kg2A_
not modelled
5.9
8
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: glutamate receptor 2;PDBTitle: ampa subtype ionotropic glutamate receptor in complex with competitive2 antagonist zk 200775
80 d1iica1
not modelled
5.8
11
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT
81 c2xzmE_
not modelled
5.8
19
PDB header: ribosomeChain: E: PDB Molecule: ribosomal protein s5 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
82 c1upiA_
not modelled
5.7
26
PDB header: epimeraseChain: A: PDB Molecule: dtdp-4-dehydrorhamnose 3,5-epimerase;PDBTitle: mycobacterium tuberculosis rmlc epimerase (rv3465)
83 c3h6hB_
not modelled
5.7
17
PDB header: membrane proteinChain: B: PDB Molecule: glutamate receptor, ionotropic kainate 2;PDBTitle: crystal structure of the glur6 amino terminal domain dimer assembly2 mpd form
84 d1zc6a1
not modelled
5.7
27
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like
85 c3zeyP_
not modelled
5.6
13
PDB header: ribosomeChain: P: PDB Molecule: 40s ribosomal protein s2, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
86 c3pz6F_
not modelled
5.6
20
PDB header: ligaseChain: F: PDB Molecule: leucyl-trna synthetase;PDBTitle: the crystal structure of glleurs-cp1
87 d2bvya1
not modelled
5.5
33
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
88 c2lmkA_
not modelled
5.5
20
PDB header: signaling proteinChain: A: PDB Molecule: exocrine gland-secreting peptide 1;PDBTitle: solution structure of mouse pheromone esp1
89 c4e1jA_
not modelled
5.5
32
PDB header: transferaseChain: A: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of glycerol kinase in complex with glycerol from2 sinorhizobium meliloti 1021
90 d2j5la1
not modelled
5.4
28
Fold: Apical membrane antigen 1Superfamily: Apical membrane antigen 1Family: Apical membrane antigen 1
91 c2j5lA_
not modelled
5.4
28
PDB header: immune systemChain: A: PDB Molecule: apical membrane antigen 1;PDBTitle: structure of a plasmodium falciparum apical membrane2 antigen 1-fab f8.12.19 complex
92 d1jb3a_
not modelled
5.4
8
Fold: OB-foldSuperfamily: TIMP-likeFamily: The laminin-binding domain of agrin
93 d1dzra_
not modelled
5.3
21
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
94 c2wfdB_
not modelled
5.3
20
PDB header: ligaseChain: B: PDB Molecule: leucyl-trna synthetase, cytoplasmic;PDBTitle: structure of the human cytosolic leucyl-trna synthetase2 editing domain
95 c1m5hF_
not modelled
5.2
23
PDB header: transferaseChain: F: PDB Molecule: formylmethanofuran--tetrahydromethanopterinPDBTitle: formylmethanofuran:tetrahydromethanopterin2 formyltransferase from archaeoglobus fulgidus
96 c2ysaA_
not modelled
5.2
28
PDB header: metal binding proteinChain: A: PDB Molecule: retinoblastoma-binding protein 6;PDBTitle: solution structure of the zinc finger cchc domain from the2 human retinoblastoma-binding protein 6 (retinoblastoma-3 binding q protein 1, rbq-1)
97 c2fwvA_
not modelled
5.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein mtubf_01000852;PDBTitle: crystal structure of rv0813
98 c1vddC_
not modelled
5.2
13
PDB header: recombinationChain: C: PDB Molecule: recombination protein recr;PDBTitle: crystal structure of recombinational repair protein recr
99 d1gr0a2
not modelled
5.2
19
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like