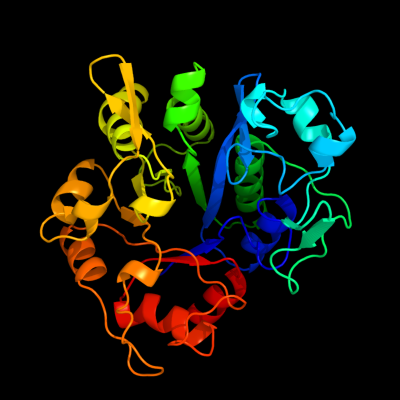
1 c1ydyA_
100.0
100
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of periplasmic glycerophosphodiester2 phosphodiesterase from escherichia coli
2 d1ydya1
100.0
100
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Glycerophosphoryl diester phosphodiesterase3 c3l12A_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: putative glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of putative glycerophosphoryl diester2 phosphodiesterase (yp_165505.1) from silicibacter pomeroyi dss-3 at3 1.60 a resolution
4 c2p76H_
100.0
33
PDB header: hydrolaseChain: H: PDB Molecule: glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of a glycerophosphodiester phosphodiesterase from2 staphylococcus aureus
5 c2pz0B_
100.0
32
PDB header: hydrolaseChain: B: PDB Molecule: glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of glycerophosphodiester phosphodiesterase (gdpd)2 from t. tengcongensis
6 c3mz2A_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of a glycerophosphoryl diester phosphodiesterase2 (bdi_3922) from parabacteroides distasonis atcc 8503 at 1.55 a3 resolution
7 c3qvqB_
100.0
26
PDB header: hydrolaseChain: B: PDB Molecule: phosphodiesterase olei02445;PDBTitle: the structure of an oleispira antarctica phosphodiesterase olei024452 in complex with the product sn-glycerol-3-phosphate
8 c2otdC_
100.0
19
PDB header: hydrolaseChain: C: PDB Molecule: glycerophosphodiester phosphodiesterase;PDBTitle: the crystal structure of the glycerophosphodiester phosphodiesterase2 from shigella flexneri 2a
9 d1zcca1
100.0
26
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Glycerophosphoryl diester phosphodiesterase10 c3ch0A_
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphodiester phosphodiesterase;PDBTitle: crystal structure of glycerophosphoryl diester phosphodiesterase2 (yp_677622.1) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
11 c3no3A_
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphodiester phosphodiesterase;PDBTitle: crystal structure of a glycerophosphodiester phosphodiesterase2 (bdi_0402) from parabacteroides distasonis atcc 8503 at 1.89 a3 resolution
12 c3ks6A_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of putative glycerophosphoryl diester2 phosphodiesterase (17743486) from agrobacterium tumefaciens3 str. c58 (dupont) at 1.80 a resolution
13 d1vd6a1
100.0
21
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Glycerophosphoryl diester phosphodiesterase14 c3i10A_
100.0
25
PDB header: hydrolaseChain: A: PDB Molecule: putative glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of putative glycerophosphoryl diester2 phosphodiesterase (np_812074.1) from bacteroides thetaiotaomicron3 vpi-5482 at 1.35 a resolution
15 c2o55A_
100.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative glycerophosphodiester phosphodiesterase;PDBTitle: crystal structure of a putative glycerophosphodiester2 phosphodiesterase from galdieria sulphuraria
16 d1o1za_
100.0
27
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Glycerophosphoryl diester phosphodiesterase17 c3rlhA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelin phosphodiesterase d lisictox-alphaia1a;PDBTitle: crystal structure of a class ii phospholipase d from loxosceles2 intermedia venom
18 c3rlgA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelin phosphodiesterase d lisictox-alphaia1a;PDBTitle: crystal structure of loxosceles intermedia phospholipase d isoform 12 h12a mutant
19 c2f9rC_
100.0
16
PDB header: hydrolaseChain: C: PDB Molecule: sphingomyelinase d 1;PDBTitle: crystal structure of the inactive state of the smase i, a2 sphingomyelinase d from loxosceles laeta venom
20 c1djyB_
98.0
16
PDB header: lipid degradationChain: B: PDB Molecule: phosphoinositide-specific phospholipase c,PDBTitle: phosphoinositide-specific phospholipase c-delta1 from rat2 complexed with inositol-2,4,5-trisphosphate
21 d1qasa3
not modelled
97.9
16
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Mammalian PLC22 d2zkmx4
not modelled
97.9
23
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Mammalian PLC23 c3ohmB_
not modelled
97.8
17
PDB header: signaling protein / hydrolaseChain: B: PDB Molecule: 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterasePDBTitle: crystal structure of activated g alpha q bound to its effector2 phospholipase c beta 3
24 c3qr0A_
not modelled
97.8
16
PDB header: hydrolaseChain: A: PDB Molecule: phospholipase c-beta (plc-beta);PDBTitle: crystal structure of s. officinalis plc21
25 c2fjuB_
not modelled
97.2
13
PDB header: signaling protein,apoptosis/hydrolaseChain: B: PDB Molecule: 1-phosphatidylinositol-4,5-bisphosphatePDBTitle: activated rac1 bound to its effector phospholipase c beta 2
26 d1vkfa_
not modelled
93.7
17
Fold: TIM beta/alpha-barrelSuperfamily: GlpP-likeFamily: GlpP-like27 c3ktsA_
not modelled
93.0
12
PDB header: transcriptional regulatorChain: A: PDB Molecule: glycerol uptake operon antiterminator regulatory protein;PDBTitle: crystal structure of glycerol uptake operon antiterminator regulatory2 protein from listeria monocytogenes str. 4b f2365
28 c2zq0B_
not modelled
92.6
6
PDB header: hydrolaseChain: B: PDB Molecule: alpha-glucosidase (alpha-glucosidase susb);PDBTitle: crystal structure of susb complexed with acarbose
29 d2gjpa2
not modelled
92.3
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain30 d1ob0a2
not modelled
90.3
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain31 c3a24A_
not modelled
89.4
17
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of bt1871 retaining glycosidase
32 d1pkla2
not modelled
88.8
13
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase33 c1mwoA_
not modelled
88.6
13
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase;PDBTitle: crystal structure analysis of the hyperthermostable2 pyrocoocus woesei alpha-amylase
34 d1e43a2
not modelled
87.0
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain35 c2qpuB_
not modelled
85.1
10
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amylase type a isozyme;PDBTitle: sugar tongs mutant s378p in complex with acarbose
36 d1e0ta2
not modelled
83.2
22
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase37 c1bplA_
not modelled
83.0
13
PDB header: glycosyltransferaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: glycosyltransferase
38 c3h4wA_
not modelled
82.5
16
PDB header: hydrolaseChain: A: PDB Molecule: phosphatidylinositol-specific phospholipase c1;PDBTitle: structure of a ca+2 dependent phosphatidylinositol-specific2 phospholipase c (pi-plc) enzyme from streptomyces antibioticus
39 c1gcyA_
not modelled
81.3
12
PDB header: hydrolaseChain: A: PDB Molecule: glucan 1,4-alpha-maltotetrahydrolase;PDBTitle: high resolution crystal structure of maltotetraose-forming2 exo-amylase
40 c3nvtA_
80.4
24
PDB header: transferase/isomeraseChain: A: PDB Molecule: 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
41 c3hvbB_
not modelled
79.3
14
PDB header: hydrolaseChain: B: PDB Molecule: protein fimx;PDBTitle: crystal structure of the dual-domain ggdef-eal module of2 fimx from pseudomonas aeruginosa
42 c2r6oB_
not modelled
78.7
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative diguanylate cyclase/phosphodiesterase (ggdef & ealPDBTitle: crystal structure of putative diguanylate cyclase/phosphodiesterase2 from thiobacillus denitrificans
43 c1jdaA_
not modelled
78.6
9
PDB header: hydrolaseChain: A: PDB Molecule: 1,4-alpha maltotetrahydrolase;PDBTitle: maltotetraose-forming exo-amylase
44 d1mxga2
not modelled
77.9
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain45 d1ht6a2
not modelled
77.7
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain46 c3thaB_
not modelled
77.0
11
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
47 c3uk2B_
not modelled
76.5
17
PDB header: ligaseChain: B: PDB Molecule: pantothenate synthetase;PDBTitle: the structure of pantothenate synthetase from burkholderia2 thailandensis
48 d1v8fa_
not modelled
75.3
20
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)49 d1avaa2
not modelled
75.1
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain50 d1ud2a2
not modelled
75.0
8
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain51 c1uasA_
not modelled
74.5
14
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of rice alpha-galactosidase
52 d1gcya2
not modelled
72.6
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain53 c3pjwA_
not modelled
72.3
16
PDB header: lyaseChain: A: PDB Molecule: cyclic dimeric gmp binding protein;PDBTitle: structure of pseudomonas fluorescence lapd ggdef-eal dual domain, i23
54 c3khdC_
not modelled
72.1
16
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pff1300w.
55 c2ejcA_
not modelled
72.0
20
PDB header: ligaseChain: A: PDB Molecule: pantoate--beta-alanine ligase;PDBTitle: crystal structure of pantoate--beta-alanine ligase (panc)2 from thermotoga maritima
56 c2qjhH_
not modelled
71.0
10
PDB header: lyaseChain: H: PDB Molecule: putative aldolase mj0400;PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
57 c2p10D_
not modelled
70.8
26
PDB header: hydrolaseChain: D: PDB Molecule: mll9387 protein;PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
58 d2d3na2
not modelled
70.4
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain59 c3guzB_
not modelled
67.6
19
PDB header: ligaseChain: B: PDB Molecule: pantothenate synthetase;PDBTitle: structural and substrate-binding studies of pantothenate2 synthenate (ps)provide insights into homotropic inhibition3 by pantoate in ps's
60 d1hvxa2
not modelled
67.2
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain61 c3hv9A_
not modelled
65.7
14
PDB header: hydrolaseChain: A: PDB Molecule: protein fimx;PDBTitle: crystal structure of fimx eal domain from pseudomonas aeruginosa
62 d2p10a1
not modelled
65.5
24
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Mll9387-like63 c3s83A_
not modelled
65.3
22
PDB header: signaling proteinChain: A: PDB Molecule: ggdef family protein;PDBTitle: crystal structure of eal domain from caulobacter crescentus cb15
64 d1h7na_
not modelled
64.6
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)65 c3n8hA_
not modelled
64.5
17
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from francisella2 tularensis
66 c1aqfB_
not modelled
58.2
16
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: pyruvate kinase from rabbit muscle with mg, k, and l-2 phospholactate
67 c2dh3A_
not modelled
57.8
24
PDB header: transport protein, signaling proteinChain: A: PDB Molecule: 4f2 cell-surface antigen heavy chain;PDBTitle: crystal structure of human ed-4f2hc
68 c1t5aB_
not modelled
54.9
20
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase, m2 isozyme;PDBTitle: human pyruvate kinase m2
69 c3blpX_
not modelled
54.5
9
PDB header: hydrolaseChain: X: PDB Molecule: alpha-amylase 1;PDBTitle: role of aromatic residues in human salivary alpha-amylase
70 c3gndC_
not modelled
54.3
16
PDB header: lyaseChain: C: PDB Molecule: aldolase lsrf;PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
71 d1ktba2
not modelled
53.9
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain72 c3gfzB_
not modelled
53.8
11
PDB header: hydrolase, signaling proteinChain: B: PDB Molecule: klebsiella pneumoniae blrp1;PDBTitle: klebsiella pneumoniae blrp1 ph 6 manganese/cy-digmp complex
73 c3pfmA_
not modelled
53.5
15
PDB header: signaling proteinChain: A: PDB Molecule: ggdef domain protein;PDBTitle: crystal structure of a eal domain of ggdef domain protein from2 pseudomonas fluorescens pf
74 d1ujpa_
not modelled
53.4
26
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes75 d2f06a2
not modelled
53.3
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like76 c2vgbB_
not modelled
53.1
19
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase isozymes r/l;PDBTitle: human erythrocyte pyruvate kinase
77 d1ua7a2
not modelled
52.6
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain78 d1pv8a_
not modelled
51.0
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)79 d1jaea2
not modelled
51.0
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain80 c2ekcA_
not modelled
50.6
23
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
81 c1pklB_
not modelled
50.2
15
PDB header: transferaseChain: B: PDB Molecule: protein (pyruvate kinase);PDBTitle: the structure of leishmania pyruvate kinase
82 d1hx0a2
not modelled
49.5
9
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain83 c3kzpA_
not modelled
48.6
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative diguanylate cyclase/phosphodiesterase;PDBTitle: crystal structure of putative diguanylate cyclase/phosphodiesterase2 from listaria monocytigenes
84 c3ma8A_
not modelled
48.3
15
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of cgd1_2040, a pyruvate kinase from cryptosporidium2 parvum
85 d1ihoa_
not modelled
47.9
19
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)86 c3e0vB_
not modelled
47.4
15
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pyruvate kinase from leishmania mexicana in2 complex with sulphate ions
87 d1bxba_
not modelled
47.1
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase88 d1rd5a_
not modelled
46.8
21
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes89 c3mxtA_
not modelled
46.5
15
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from campylobacter2 jejuni
90 d1r46a2
not modelled
45.0
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain91 d1qopa_
not modelled
43.3
21
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes92 d1kjqa2
not modelled
42.4
8
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like93 c3l2iB_
not modelled
42.0
19
PDB header: lyaseChain: B: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
94 c3navB_
not modelled
41.7
21
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
95 d2f6ua1
not modelled
41.3
14
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases96 d2g50a2
not modelled
41.2
16
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase97 c1ud8A_
not modelled
41.1
6
PDB header: hydrolaseChain: A: PDB Molecule: amylase;PDBTitle: crystal structure of amyk38 with lithium ion
98 d1tqxa_
not modelled
41.1
21
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase99 d1a3xa2
not modelled
40.4
13
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase100 c3t07D_
not modelled
40.4
15
PDB header: transferase/transferase inhibitorChain: D: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
101 c3femB_
not modelled
40.0
24
PDB header: biosynthetic protein, transferaseChain: B: PDB Molecule: pyridoxine biosynthesis protein snz1;PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
102 c3obkH_
not modelled
40.0
16
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
103 c2nv2U_
not modelled
39.8
27
PDB header: lyase/transferaseChain: U: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: structure of the plp synthase complex pdx1/2 (yaad/e) from bacillus2 subtilis
104 d2ptda_
not modelled
39.2
21
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Bacterial PLC105 c3o6cA_
not modelled
39.0
13
PDB header: transferaseChain: A: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: pyridoxal phosphate biosynthetic protein pdxj from campylobacter2 jejuni
106 c2zbtB_
not modelled
38.9
20
PDB header: lyaseChain: B: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: crystal structure of pyridoxine biosynthesis protein from thermus2 thermophilus hb8
107 c2htmB_
not modelled
38.3
14
PDB header: biosynthetic proteinChain: B: PDB Molecule: thiazole biosynthesis protein thig;PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
108 c1a3wB_
not modelled
38.2
12
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: pyruvate kinase from saccharomyces cerevisiae complexed with fbp, pg,2 mn2+ and k+
109 c2e28A_
not modelled
38.2
16
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
110 d2basa1
not modelled
37.8
10
Fold: TIM beta/alpha-barrelSuperfamily: EAL domain-likeFamily: EAL domain111 c1kjjA_
not modelled
37.7
8
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase 2;PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
112 d1ojxa_
not modelled
36.4
12
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase113 c1jaeA_
not modelled
36.0
13
PDB header: glycosidaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: structure of tenebrio molitor larval alpha-amylase
114 c3a5vA_
not modelled
35.0
16
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of alpha-galactosidase i from mortierella vinacea
115 c2w27A_
not modelled
33.7
11
PDB header: signaling proteinChain: A: PDB Molecule: ykui protein;PDBTitle: crystal structure of the bacillus subtilis ykui protein,2 with an eal domain, in complex with substrate c-di-gmp and3 calcium
116 d3dhpa2
not modelled
33.4
8
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain117 d1gzga_
not modelled
32.7
18
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)118 c1e0tD_
not modelled
32.6
17
PDB header: phosphotransferaseChain: D: PDB Molecule: pyruvate kinase;PDBTitle: r292d mutant of e. coli pyruvate kinase
119 d1uasa2
not modelled
32.5
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain120 c3eoeC_
not modelled
32.4
15
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007