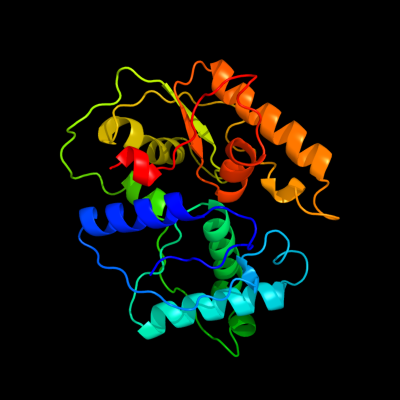
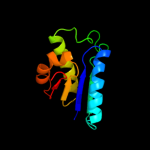
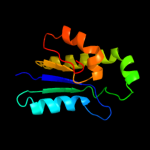
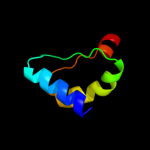
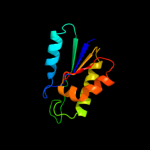
1 c3kwlA_
99.9
11
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical protein from helicobacter pylori
2 c3ojcD_
92.0
14
PDB header: isomeraseChain: D: PDB Molecule: putative aspartate/glutamate racemase;PDBTitle: crystal structure of a putative asp/glu racemase from yersinia pestis
3 c3s81A_
91.0
9
PDB header: isomeraseChain: A: PDB Molecule: putative aspartate racemase;PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
4 c2dx7B_
90.1
18
PDB header: isomeraseChain: B: PDB Molecule: aspartate racemase;PDBTitle: crystal structure of pyrococcus horikoshii ot3 aspartate racemase2 complex with citric acid
5 c3outC_
89.9
12
PDB header: isomeraseChain: C: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
6 c2zskA_
87.1
15
PDB header: unknown functionChain: A: PDB Molecule: 226aa long hypothetical aspartate racemase;PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
7 c3hfrA_
78.4
13
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
8 c1b74A_
78.4
15
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: glutamate racemase from aquifex pyrophilus
9 c2jfoB_
77.9
18
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
10 d1jfla1
77.1
10
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase11 c2dwuA_
76.1
14
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
12 c2jfqA_
73.9
15
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of staphylococcus aureus glutamate2 racemase in complex with d-glutamate
13 c2jfzB_
73.7
17
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
14 c2gzmB_
70.1
14
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of the glutamate racemase from bacillus2 anthracis
15 c2ohoA_
69.3
11
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: structural basis for glutamate racemase inhibitor
16 c1zuwA_
66.9
12
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase 1;PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
17 d1vk8a_
64.4
13
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like18 d1b74a1
53.6
15
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase19 c2eq5D_
50.4
6
PDB header: isomeraseChain: D: PDB Molecule: 228aa long hypothetical hydantoin racemase;PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
20 d3c8ya1
50.2
15
Fold: Fe-only hydrogenaseSuperfamily: Fe-only hydrogenaseFamily: Fe-only hydrogenase21 c2epiA_
not modelled
49.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0045 protein mj1052;PDBTitle: crystal structure pf hypothetical protein mj1052 from2 methanocaldococcus jannascii (form 2)
22 c1c4cA_
not modelled
46.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (fe-only hydrogenase);PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
23 c3lx4B_
not modelled
45.2
14
PDB header: oxidoreductaseChain: B: PDB Molecule: fe-hydrogenase;PDBTitle: stepwise [fefe]-hydrogenase h-cluster assembly revealed in the2 structure of hyda(deltaefg)
24 d1lxja_
not modelled
44.1
7
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like25 c3hlyA_
not modelled
42.3
16
PDB header: flavoproteinChain: A: PDB Molecule: flavodoxin-like domain;PDBTitle: crystal structure of the flavodoxin-like domain from2 synechococcus sp q5mzp6_synp6 protein. northeast structural3 genomics consortium target snr135d.
26 d1hfel1
not modelled
40.9
11
Fold: Fe-only hydrogenaseSuperfamily: Fe-only hydrogenaseFamily: Fe-only hydrogenase27 d2a9va1
not modelled
39.1
15
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)28 c3qvjB_
not modelled
38.8
10
PDB header: isomeraseChain: B: PDB Molecule: putative hydantoin racemase;PDBTitle: allantoin racemase from klebsiella pneumoniae
29 d2iboa1
not modelled
38.7
9
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like30 d1jfla2
not modelled
37.8
17
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase31 c3cf4A_
not modelled
37.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: acetyl-coa decarboxylase/synthase alpha subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex
32 c2jfnA_
not modelled
35.4
12
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
33 c1hfeL_
not modelled
34.3
11
PDB header: hydrogenaseChain: L: PDB Molecule: protein (fe-only hydrogenase (e.c.1.18.99.1)PDBTitle: 1.6 a resolution structure of the fe-only hydrogenase from2 desulfovibrio desulfuricans
34 d1yqha1
not modelled
32.3
11
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like35 d2fzva1
not modelled
31.5
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase36 c1gx7A_
not modelled
29.4
11
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic [fe] hydrogenase large subunit;PDBTitle: best model of the electron transfer complex between2 cytochrome c3 and [fe]-hydrogenase
37 c1x0lB_
not modelled
25.5
19
PDB header: oxidoreductaseChain: B: PDB Molecule: homoisocitrate dehydrogenase;PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
38 d3pmga1
not modelled
24.9
16
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains39 c3fniA_
not modelled
22.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
40 d2dx7a1
not modelled
21.3
17
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase41 d1xaca_
not modelled
20.9
24
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases42 c2ct6A_
not modelled
20.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 domain-binding glutamic acid-rich-likePDBTitle: solution structure of the sh3 domain-binding glutamic acid-2 rich-like protein 2
43 c1u6tA_
not modelled
20.2
12
PDB header: protein binding, signaling proteinChain: A: PDB Molecule: sh3 domain-binding glutamic acid-rich-likePDBTitle: crystal structure of the human sh3 binding glutamic-rich2 protein like
44 d1lxna_
not modelled
19.8
16
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like45 d1kfia1
not modelled
19.1
16
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains46 c2fb6A_
not modelled
17.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: structure of conserved protein of unknown function bt1422 from2 bacteroides thetaiotaomicron
47 c3blxL_
not modelled
17.2
13
PDB header: oxidoreductaseChain: L: PDB Molecule: isocitrate dehydrogenase [nad] subunit 2;PDBTitle: yeast isocitrate dehydrogenase (apo form)
48 d1z2wa1
not modelled
16.2
13
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: YfcE-like49 c3blxM_
not modelled
15.5
13
PDB header: oxidoreductaseChain: M: PDB Molecule: isocitrate dehydrogenase [nad] subunit 1;PDBTitle: yeast isocitrate dehydrogenase (apo form)
50 c3rh0A_
not modelled
15.3
22
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum mycothiol/mycoredoxin1-dependent arsenate2 reductase cg_arsc2
51 d1rtta_
not modelled
15.3
10
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase52 d2auna2
not modelled
15.3
12
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like53 c3l3bA_
not modelled
14.0
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: es1 family protein;PDBTitle: crystal structure of isoprenoid biosynthesis protein with2 amidotransferase-like domain from ehrlichia chaffeensis at 1.90a3 resolution
54 c3ijfX_
not modelled
13.5
11
PDB header: hydrolaseChain: X: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase from mycobacterium2 tuberculosis
55 c3h75A_
not modelled
13.1
11
PDB header: sugar binding proteinChain: A: PDB Molecule: periplasmic sugar-binding domain protein;PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
56 c3onoA_
not modelled
12.3
17
PDB header: isomeraseChain: A: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
57 d1j09a2
not modelled
11.9
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain58 c3gjzB_
not modelled
11.7
18
PDB header: immune systemChain: B: PDB Molecule: microcin immunity protein mccf;PDBTitle: crystal structure of microcin immunity protein mccf from bacillus2 anthracis str. ames
59 d2bisa1
not modelled
11.7
20
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Glycosyl transferases group 160 c3c8nB_
not modelled
11.3
8
PDB header: oxidoreductaseChain: B: PDB Molecule: probable f420-dependent glucose-6-phosphate dehydrogenasePDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
61 c2jr7A_
not modelled
11.0
23
PDB header: metal binding proteinChain: A: PDB Molecule: dph3 homolog;PDBTitle: solution structure of human desr1
62 d1w0da_
not modelled
11.0
16
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases63 c1c4gB_
not modelled
10.6
16
PDB header: transferaseChain: B: PDB Molecule: protein (alpha-d-glucose 1-phosphatePDBTitle: phosphoglucomutase vanadate based transition state analog2 complex
64 c2dgdD_
not modelled
10.0
10
PDB header: lyaseChain: D: PDB Molecule: 223aa long hypothetical arylmalonate decarboxylase;PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
65 d1wgea1
not modelled
9.9
19
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger66 c3o1hB_
not modelled
9.9
19
PDB header: signaling proteinChain: B: PDB Molecule: periplasmic protein tort;PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
67 d1fjha_
not modelled
9.9
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases68 c2vlbC_
not modelled
9.6
13
PDB header: lyaseChain: C: PDB Molecule: arylmalonate decarboxylase;PDBTitle: structure of unliganded arylmalonate decarboxylase
69 c1zrsB_
not modelled
9.5
14
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protein;PDBTitle: wild-type ld-carboxypeptidase
70 c2fzvC_
not modelled
9.5
8
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: putative arsenical resistance protein;PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
71 d1h75a_
not modelled
9.3
10
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase72 c3afhA_
not modelled
9.1
12
PDB header: ligaseChain: A: PDB Molecule: glutamyl-trna synthetase 2;PDBTitle: crystal structure of thermotoga maritima nondiscriminating glutamyl-2 trna synthetase in complex with a glutamyl-amp analog
73 c3u1hA_
not modelled
9.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-isopropylmalate dehydrogenase;PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
74 d1rbli_
not modelled
9.1
7
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit75 c2wlbB_
not modelled
8.3
13
PDB header: electron transportChain: B: PDB Molecule: electron transfer protein 1, mitochondrial;PDBTitle: adrenodoxin-like ferredoxin etp1fd(516-618) of2 schizosaccharomyces pombe mitochondria
76 c3lcmB_
not modelled
8.2
9
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
77 c2z0fA_
not modelled
8.2
14
PDB header: isomeraseChain: A: PDB Molecule: putative phosphoglucomutase;PDBTitle: crystal structure of putative phosphoglucomutase from thermus2 thermophilus hb8
78 d1pn3a_
not modelled
8.2
15
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Gtf glycosyltransferase79 c2x49A_
not modelled
8.2
16
PDB header: protein transportChain: A: PDB Molecule: invasion protein inva;PDBTitle: crystal structure of the c-terminal domain of inva
80 c3na7A_
not modelled
8.2
12
PDB header: gene regulation, chaperoneChain: A: PDB Molecule: hp0958;PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
81 c2gejA_
not modelled
8.1
14
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol mannosyltransferase (pima);PDBTitle: crystal structure of phosphatidylinositol mannosyltransferase (pima)2 from mycobacterium smegmatis in complex with gdp-man
82 c2ppwA_
not modelled
7.6
10
PDB header: isomeraseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
83 c3h7hA_
not modelled
7.6
17
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt4;PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
84 d1r7ha_
not modelled
7.6
13
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase85 c3c4vB_
not modelled
7.6
20
PDB header: transferaseChain: B: PDB Molecule: predicted glycosyltransferases;PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
86 c3he8A_
not modelled
7.3
18
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
87 c3ikzA_
not modelled
7.3
8
PDB header: transferaseChain: A: PDB Molecule: phosphopantetheine adenylyltransferase;PDBTitle: crystal structure of phosphopantetheine adenylyltransferase from2 burkholderia pseudomallei
88 d1b43a2
not modelled
7.0
20
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain89 d1g2ua_
not modelled
7.0
21
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases90 c2fuvB_
not modelled
6.9
23
PDB header: isomeraseChain: B: PDB Molecule: phosphoglucomutase;PDBTitle: phosphoglucomutase from salmonella typhimurium.
91 c3c5yD_
not modelled
6.9
11
PDB header: isomeraseChain: D: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
92 d2d30a1
not modelled
6.9
11
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase93 d1coza_
not modelled
6.9
7
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Cytidylyltransferase94 c2jadA_
not modelled
6.9
7
PDB header: electron transportChain: A: PDB Molecule: yellow fluorescent protein glutaredoxin fusionPDBTitle: yellow fluorescent protein - glutaredoxin fusion protein
95 c3nbmA_
not modelled
6.7
18
PDB header: transferaseChain: A: PDB Molecule: pts system, lactose-specific iibc components;PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
96 c3dmoD_
not modelled
6.7
14
PDB header: hydrolaseChain: D: PDB Molecule: cytidine deaminase;PDBTitle: 1.6 a crystal structure of cytidine deaminase from2 burkholderia pseudomallei
97 c3orsD_
not modelled
6.6
13
PDB header: isomerase,biosynthetic proteinChain: D: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
98 d8abpa_
not modelled
6.5
13
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like99 c3fmfA_
not modelled
6.5
11
PDB header: ligaseChain: A: PDB Molecule: dethiobiotin synthetase;PDBTitle: crystal structure of mycobacterium tuberculosis dethiobiotin2 synthetase complexed with 7,8 diaminopelargonic acid carbamate