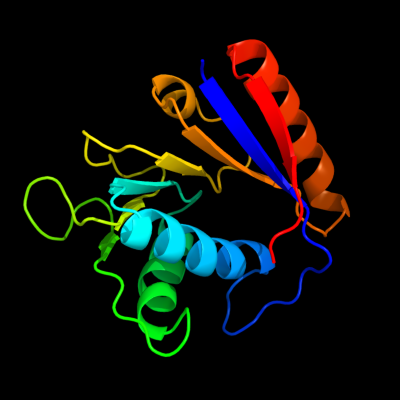
1 d1em8a_
100.0
100
Fold: DNA polymerase III chi subunitSuperfamily: DNA polymerase III chi subunitFamily: DNA polymerase III chi subunit2 d1c4oa2
80.7
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain3 c2j17A_
80.2
7
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase yil113w;PDBTitle: ptyr bound form of sdp-1
4 c2imgA_
78.7
17
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 23;PDBTitle: crystal structure of dual specificity protein phosphatase2 23 from homo sapiens in complex with ligand malate ion
5 d1tf5a4
78.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain6 d1t5la2
77.2
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain7 d1fuka_
76.2
8
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain8 d1nkta4
69.0
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain9 c3rgqA_
67.5
15
PDB header: hydrolaseChain: A: PDB Molecule: protein-tyrosine phosphatase mitochondrial 1;PDBTitle: crystal structure of ptpmt1 in complex with pi(5)p
10 c3hgtA_
62.2
13
PDB header: transcriptionChain: A: PDB Molecule: hda1 complex subunit 3;PDBTitle: structural and functional studies of the yeast class ii hda12 hdac complex
11 c1fpzF_
62.0
10
PDB header: hydrolaseChain: F: PDB Molecule: cyclin-dependent kinase inhibitor 3;PDBTitle: crystal structure analysis of kinase associated phosphatase2 (kap) with a substitution of the catalytic site cysteine3 (cys140) to a serine
12 c2p4dA_
61.3
20
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase;PDBTitle: structure-assisted discovery of variola major h12 phosphatase inhibitors
13 c1nl3B_
61.0
25
PDB header: protein transportChain: B: PDB Molecule: preprotein translocase seca 1 subunit;PDBTitle: crystal structure of the seca protein translocation atpase2 from mycobacterium tuberculosis in apo form
14 c2esbA_
57.1
15
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 18;PDBTitle: crystal structure of human dusp18
15 d1fpza_
56.5
10
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like16 d1t5ia_
53.1
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain17 c1yz4A_
52.6
10
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase-like 15 isoform a;PDBTitle: crystal structure of dusp15
18 c3dl8B_
49.5
20
PDB header: protein transportChain: B: PDB Molecule: protein translocase subunit seca;PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
19 d1gm5a4
49.2
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain20 c1tf2A_
49.1
20
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of seca:adp in an open conformation from2 bacillus subtilis
21 d2eyqa2
not modelled
48.4
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain22 c2vdaA_
not modelled
45.9
19
PDB header: protein transportChain: A: PDB Molecule: translocase subunit seca;PDBTitle: solution structure of the seca-signal peptide complex
23 c2vbcA_
not modelled
45.4
26
PDB header: hydrolaseChain: A: PDB Molecule: dengue 4 ns3 full-length protein;PDBTitle: crystal structure of the ns3 protease-helicase from dengue2 virus
24 c2hcmA_
not modelled
45.4
16
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase;PDBTitle: crystal structure of mouse putative dual specificity phosphatase2 complexed with zinc tungstate, new york structural genomics3 consortium
25 c2hk8B_
not modelled
44.5
18
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
26 d1oywa3
not modelled
44.1
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain27 c1wrmA_
not modelled
41.8
15
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase 22;PDBTitle: crystal structure of jsp-1
28 c3juxA_
not modelled
41.7
17
PDB header: protein transportChain: A: PDB Molecule: protein translocase subunit seca;PDBTitle: structure of the translocation atpase seca from thermotoga2 maritima
29 c2oudA_
not modelled
41.4
24
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of the catalytic domain of human mkp5
30 c3dinB_
not modelled
36.5
18
PDB header: membrane protein, protein transportChain: B: PDB Molecule: protein translocase subunit seca;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
31 d1jr6a_
not modelled
35.6
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RNA helicase32 c2jmkA_
not modelled
35.4
19
PDB header: protein bindingChain: A: PDB Molecule: hypothetical protein ta0956;PDBTitle: solution structure of ta0956
33 c1zzwA_
not modelled
34.7
23
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of catalytic domain of human map kinase2 phosphatase 5
34 c1p74B_
not modelled
34.6
11
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase (aroe) from2 haemophilus influenzae
35 c3dmdA_
not modelled
34.6
22
PDB header: transport proteinChain: A: PDB Molecule: signal recognition particle receptor;PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
36 c3i32A_
not modelled
34.5
14
PDB header: rna binding protein,hydrolaseChain: A: PDB Molecule: heat resistant rna dependent atpase;PDBTitle: dimeric structure of a hera helicase fragment including the c-terminal2 reca domain, the dimerization domain, and the rna binding domain
37 c1ii0A_
not modelled
33.9
25
PDB header: hydrolaseChain: A: PDB Molecule: arsenical pump-driving atpase;PDBTitle: crystal structure of the escherichia coli arsenite-translocating2 atpase
38 d1ihua1
not modelled
33.5
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like39 c2wv9A_
not modelled
32.9
18
PDB header: hydrolaseChain: A: PDB Molecule: flavivirin protease ns2b regulatory subunit, flavivirinPDBTitle: crystal structure of the ns3 protease-helicase from murray2 valley encephalitis virus
40 c2wgpA_
not modelled
32.8
9
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 14;PDBTitle: crystal structure of human dual specificity phosphatase 14
41 c2eyqA_
not modelled
32.7
13
PDB header: hydrolaseChain: A: PDB Molecule: transcription-repair coupling factor;PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
42 c2qeqA_
not modelled
31.6
20
PDB header: hydrolaseChain: A: PDB Molecule: flavivirin protease ns3 catalytic subunit;PDBTitle: crystal structure of kunjin virus ns3 helicase
43 d1m3ga_
not modelled
30.8
22
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like44 c3emuA_
not modelled
30.0
11
PDB header: hydrolaseChain: A: PDB Molecule: leucine rich repeat and phosphatase domainPDBTitle: crystal structure of a leucine rich repeat and phosphatase2 domain containing protein from entamoeba histolytica
45 d1t5la1
not modelled
29.9
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain46 d1mkpa_
not modelled
29.1
14
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like47 c2nt2C_
not modelled
28.5
13
PDB header: hydrolaseChain: C: PDB Molecule: protein phosphatase slingshot homolog 2;PDBTitle: crystal structure of slingshot phosphatase 2
48 c3nmeA_
not modelled
27.6
12
PDB header: hydrolaseChain: A: PDB Molecule: sex4 glucan phosphatase;PDBTitle: structure of a plant phosphatase
49 c2fsgA_
not modelled
26.5
21
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase seca subunit;PDBTitle: complex seca:atp from escherichia coli
50 d1xo1a2
not modelled
25.6
10
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain51 c2g6zB_
not modelled
25.3
18
PDB header: hydrolaseChain: B: PDB Molecule: dual specificity protein phosphatase 5;PDBTitle: crystal structure of human dusp5
52 c3d4oA_
not modelled
24.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: dipicolinate synthase subunit a;PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
53 c3e59A_
not modelled
24.2
11
PDB header: transferaseChain: A: PDB Molecule: pyoverdine biosynthesis protein pvca;PDBTitle: crystal structure of the pvca (pa2254) protein from pseudomonas2 aeruginosa
54 d2eyqa5
not modelled
24.0
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain55 c2j37W_
not modelled
22.7
22
PDB header: ribosomeChain: W: PDB Molecule: signal recognition particle 54 kda proteinPDBTitle: model of mammalian srp bound to 80s rncs
56 d1tfra2
not modelled
22.4
15
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain57 c1gl9B_
not modelled
21.5
13
PDB header: topoisomeraseChain: B: PDB Molecule: reverse gyrase;PDBTitle: archaeoglobus fulgidus reverse gyrase complexed with adpnp
58 c2iy3A_
not modelled
20.4
16
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein ffh;PDBTitle: structure of the e. coli signal recognition particle2 bound to a translating ribosome
59 d1fx0a3
not modelled
20.4
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)60 d2g2ja1
not modelled
20.2
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain61 c2r0bA_
not modelled
19.6
17
PDB header: hydrolaseChain: A: PDB Molecule: serine/threonine/tyrosine-interacting protein;PDBTitle: crystal structure of human tyrosine phosphatase-like2 serine/threonine/tyrosine-interacting protein
62 c3dm5A_
not modelled
19.0
19
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
63 c1rrzA_
not modelled
18.9
16
PDB header: structural genomics,biosynthetic proteinChain: A: PDB Molecule: glycogen synthesis protein glgs;PDBTitle: solution structure of glgs protein from e. coli
64 d1rrza_
not modelled
18.9
16
Fold: Spectrin repeat-likeSuperfamily: Glycogen synthesis protein GlgSFamily: Glycogen synthesis protein GlgS65 d2jdia3
not modelled
18.1
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)66 c2z83A_
not modelled
17.6
18
PDB header: viral proteinChain: A: PDB Molecule: helicase/nucleoside triphosphatase;PDBTitle: crystal structure of catalytic domain of japanese2 encephalitis virus ns3 helicase/nucleoside triphosphatase3 at a resolution 1.8
67 d1z5za1
not modelled
17.5
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain68 d2p6ra4
not modelled
17.1
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain69 c2cnwF_
not modelled
17.0
16
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
70 d1cmwa2
not modelled
16.5
16
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain71 d1j8yf2
not modelled
16.4
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like72 c1gm5A_
not modelled
15.6
14
PDB header: helicaseChain: A: PDB Molecule: recg;PDBTitle: structure of recg bound to three-way dna junction
73 d2j0sa2
not modelled
14.9
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain74 d2a90a1
not modelled
14.9
5
Fold: WWE domainSuperfamily: WWE domainFamily: WWE domain75 c2x0dA_
not modelled
14.7
7
PDB header: transferaseChain: A: PDB Molecule: wsaf;PDBTitle: apo structure of wsaf
76 d1pjca1
not modelled
14.7
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain77 d1z3ix1
not modelled
13.8
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain78 c1zu4A_
not modelled
13.6
25
PDB header: protein transportChain: A: PDB Molecule: ftsy;PDBTitle: crystal structure of ftsy from mycoplasma mycoides- space2 group p21212
79 c1vmaA_
not modelled
13.5
22
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
80 c2ihnA_
not modelled
13.5
15
PDB header: hydrolase/dnaChain: A: PDB Molecule: ribonuclease h;PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
81 c2pqmA_
not modelled
13.5
18
PDB header: lyaseChain: A: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of cysteine synthase (oass) from entamoeba2 histolytica at 1.86 a resolution
82 c2j7pA_
not modelled
13.4
16
PDB header: signal recognitionChain: A: PDB Molecule: signal recognition particle protein;PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
83 d1hv8a2
not modelled
13.3
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain84 c3ia7A_
not modelled
13.0
14
PDB header: transferaseChain: A: PDB Molecule: calg4;PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
85 c2hjvB_
not modelled
12.7
14
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase dbpa;PDBTitle: structure of the second domain (residues 207-368) of the2 bacillus subtilis yxin protein
86 c2ipcB_
not modelled
12.3
25
PDB header: transport proteinChain: B: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of the translocation atpase seca from thermus2 thermophilus reveals a parallel, head-to-head dimer
87 c1j8yF_
not modelled
12.1
16
PDB header: signaling proteinChain: F: PDB Molecule: signal recognition 54 kda protein;PDBTitle: signal recognition particle conserved gtpase domain from a.2 ambivalens t112a mutant
88 d1b74a1
not modelled
12.0
16
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase89 c2v6jA_
not modelled
11.9
24
PDB header: hydrolaseChain: A: PDB Molecule: rna helicase;PDBTitle: kokobera virus helicase: mutant met47thr
90 d1skyb3
not modelled
11.8
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)91 d2b2na1
not modelled
11.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain92 d2pd4a1
not modelled
11.6
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases93 c3bxzA_
not modelled
11.4
18
PDB header: transport proteinChain: A: PDB Molecule: preprotein translocase subunit seca;PDBTitle: crystal structure of the isolated dead motor domains from2 escherichia coli seca
94 c1zq1B_
not modelled
11.0
28
PDB header: lyaseChain: B: PDB Molecule: glutamyl-trna(gln) amidotransferase subunit d;PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
95 c2qsdB_
not modelled
11.0
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of a protein il1583 from idiomarina loihiensis
96 d1t6t1_
not modelled
10.8
9
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain97 c3u62A_
not modelled
10.6
14
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase from thermotoga maritima
98 c3b9qA_
not modelled
10.4
25
PDB header: protein transportChain: A: PDB Molecule: chloroplast srp receptor homolog, alpha subunitPDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
99 c3fshC_
not modelled
10.1
55
PDB header: ligaseChain: C: PDB Molecule: autocrine motility factor receptor, isoform 2;PDBTitle: crystal structure of the ubiquitin conjugating enzyme2 ube2g2 bound to the g2br domain of ubiquitin ligase gp78