1 c1h0mD_
100.0
19
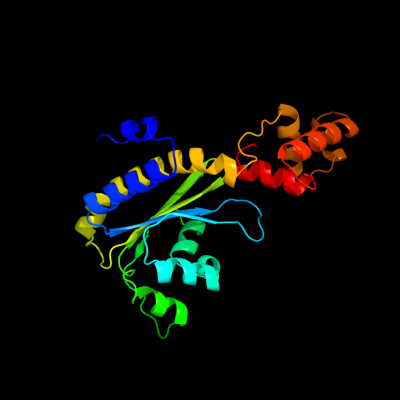
PDB header: transcription/dnaChain: D: PDB Molecule: transcriptional activator protein trar;PDBTitle: three-dimensional structure of the quorum sensing protein2 trar bound to its autoinducer and to its target dna
2 c2q0oA_
100.0
22
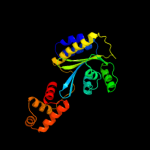
PDB header: transcriptionChain: A: PDB Molecule: probable transcriptional activator protein trar;PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
3 c3sztB_
100.0
31
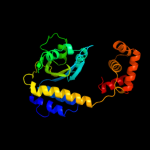
PDB header: transcriptionChain: B: PDB Molecule: quorum-sensing control repressor;PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
4 c3qp5C_
100.0
26
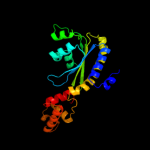
PDB header: transcriptionChain: C: PDB Molecule: cvir transcriptional regulator;PDBTitle: crystal structure of cvir bound to antagonist chlorolactone (cl)
5 c2avxA_
100.0
99
PDB header: transcriptionChain: A: PDB Molecule: regulatory protein sdia;PDBTitle: solution structure of e coli sdia1-171
6 d1l3la2
99.8
16
Fold: Profilin-likeSuperfamily: Pheromone-binding domain of LuxR-like quorum-sensing transcription factorsFamily: Pheromone-binding domain of LuxR-like quorum-sensing transcription factors7 c2uv0G_
99.8
20
PDB header: transcriptionChain: G: PDB Molecule: transcriptional activator protein lasr;PDBTitle: structure of the p. aeruginosa lasr ligand-binding domain2 bound to its autoinducer
8 c3qp1A_
99.8
20
PDB header: transcriptionChain: A: PDB Molecule: cvir transcriptional regulator;PDBTitle: crystal structure of cvir ligand-binding domain bound to the native2 ligand c6-hsl
9 c3cloC_
99.8
41
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator containing a2 luxr dna binding domain (np_811094.1) from bacteroides3 thetaiotaomicron vpi-5482 at 2.04 a resolution
10 c2krfB_
99.7
23
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein coma;PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
11 d1l3la1
99.7
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)12 c1zljE_
99.7
25
PDB header: transcriptionChain: E: PDB Molecule: dormancy survival regulator;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
13 d1p4wa_
99.7
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)14 d1fsea_
99.7
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)15 d1a04a1
99.7
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)16 c2rnjA_
99.7
36
PDB header: transcriptionChain: A: PDB Molecule: response regulator protein vrar;PDBTitle: nmr structure of the s. aureus vrar dna binding domain
17 c3c3wB_
99.7
25
PDB header: transcriptionChain: B: PDB Molecule: two component transcriptional regulatory protein devr;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
18 c1x3uA_
99.7
31
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein fixj;PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
19 c3klnC_
99.7
33
PDB header: transcriptionChain: C: PDB Molecule: transcriptional regulator, luxr family;PDBTitle: vibrio cholerae vpst
20 c1rnlA_
99.6
31
PDB header: signal transduction proteinChain: A: PDB Molecule: nitrate/nitrite response regulator protein narl;PDBTitle: the nitrate/nitrite response regulator protein narl from narl
21 c2jpcA_
not modelled
99.6
34
PDB header: dna binding proteinChain: A: PDB Molecule: ssrb;PDBTitle: ssrb dna binding protein
22 d1yioa1
not modelled
99.6
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)23 c1zn2A_
not modelled
99.6
27
PDB header: transcription regulatorChain: A: PDB Molecule: response regulatory protein;PDBTitle: low resolution structure of response regulator styr
24 d1ttya_
not modelled
98.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain25 c1kgsA_
not modelled
98.3
25
PDB header: dna binding proteinChain: A: PDB Molecule: dna binding response regulator d;PDBTitle: crystal structure at 1.50 a of an ompr/phob homolog from thermotoga2 maritima
26 d2p7vb1
not modelled
98.3
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain27 d1smyf2
not modelled
98.2
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain28 d1rp3a2
not modelled
98.2
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain29 d1xsva_
not modelled
98.2
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like30 d1ku7a_
not modelled
98.2
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain31 c2o8xA_
not modelled
98.2
14
PDB header: transcriptionChain: A: PDB Molecule: probable rna polymerase sigma-c factor;PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
32 d1or7a1
not modelled
98.1
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain33 c3hugA_
not modelled
98.1
23
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
34 d1ku3a_
not modelled
98.1
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain35 d1s7oa_
not modelled
98.0
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like36 c3mzyA_
not modelled
98.0
37
PDB header: rna binding proteinChain: A: PDB Molecule: rna polymerase sigma-h factor;PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
37 c3q9sA_
not modelled
98.0
25
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding response regulator;PDBTitle: crystal structure of rra(1-215) from deinococcus radiodurans
38 c1rp3G_
not modelled
98.0
23
PDB header: transcriptionChain: G: PDB Molecule: rna polymerase sigma factor sigma-28 (flia);PDBTitle: cocrystal structure of the flagellar sigma/anti-sigma2 complex, sigma-28/flgm
39 c1or7A_
not modelled
97.9
19
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-e factor;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
40 c2q1zA_
not modelled
97.7
9
PDB header: transcriptionChain: A: PDB Molecule: rpoe, ecf sige;PDBTitle: crystal structure of rhodobacter sphaeroides sige in complex with the2 anti-sigma chrr
41 c3t72o_
not modelled
97.5
16
PDB header: transcription/dnaChain: O: PDB Molecule: pho box dna (strand 1);PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
42 c2oqrA_
not modelled
97.5
19
PDB header: transcription,signaling proteinChain: A: PDB Molecule: sensory transduction protein regx3;PDBTitle: the structure of the response regulator regx3 from mycobacterium2 tuberculosis
43 c1l9uH_
not modelled
97.4
16
PDB header: transcriptionChain: H: PDB Molecule: sigma factor siga;PDBTitle: thermus aquaticus rna polymerase holoenzyme at 4 a2 resolution
44 c2a6eF_
not modelled
97.3
14
PDB header: transferaseChain: F: PDB Molecule: rna polymerase sigma factor rpod;PDBTitle: crystal structure of the t. thermophilus rna polymerase2 holoenzyme
45 d1jhfa1
not modelled
97.2
30
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain46 c2hqrA_
not modelled
97.1
13
PDB header: signaling proteinChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: structure of a atypical orphan response regulator protein revealed a2 new phosphorylation-independent regulatory mechanism
47 d2cg4a1
not modelled
97.0
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain48 c1ys7B_
not modelled
96.9
25
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulatory protein prra;PDBTitle: crystal structure of the response regulator protein prra comlexed with2 mg2+
49 d1stza1
not modelled
96.8
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain50 c2gwrA_
not modelled
96.8
16
PDB header: signaling proteinChain: A: PDB Molecule: dna-binding response regulator mtra;PDBTitle: crystal structure of the response regulator protein mtra from2 mycobacterium tuberculosis
51 c3p7nB_
not modelled
96.6
28
PDB header: dna binding proteinChain: B: PDB Molecule: sensor histidine kinase;PDBTitle: crystal structure of light activated transcription factor el222 from2 erythrobacter litoralis
52 c3k2zA_
not modelled
96.5
23
PDB header: hydrolaseChain: A: PDB Molecule: lexa repressor;PDBTitle: crystal structure of a lexa protein from thermotoga maritima
53 c1u78A_
not modelled
96.5
14
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
54 c2wteB_
not modelled
96.4
27
PDB header: antiviral proteinChain: B: PDB Molecule: csa3;PDBTitle: the structure of the crispr-associated protein, csa3, from2 sulfolobus solfataricus at 1.8 angstrom resolution.
55 c3r0jA_
not modelled
96.4
22
PDB header: dna binding proteinChain: A: PDB Molecule: possible two component system response transcriptionalPDBTitle: structure of phop from mycobacterium tuberculosis
56 d1biaa1
not modelled
96.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like57 c2l4aA_
not modelled
96.3
9
PDB header: dna binding proteinChain: A: PDB Molecule: leucine responsive regulatory protein;PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
58 d2cyya1
not modelled
96.3
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain59 d1i1ga1
not modelled
96.3
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain60 d2d1ha1
not modelled
96.3
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: TrmB-like61 d2cfxa1
not modelled
96.3
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain62 c1p2fA_
not modelled
96.1
28
PDB header: transcriptionChain: A: PDB Molecule: response regulator;PDBTitle: crystal structure analysis of response regulator drrb, a2 thermotoga maritima ompr/phob homolog
63 c2cg4B_
not modelled
96.1
19
PDB header: transcriptionChain: B: PDB Molecule: regulatory protein asnc;PDBTitle: structure of e.coli asnc
64 c2e1cA_
not modelled
96.1
19
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
65 d1j5ya1
not modelled
96.1
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like66 c3iydF_
not modelled
96.1
16
PDB header: transcription/dnaChain: F: PDB Molecule: rna polymerase sigma factor rpod;PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
67 c2dbbA_
not modelled
96.0
16
PDB header: transcriptional regulatorChain: A: PDB Molecule: putative hth-type transcriptional regulator ph0061;PDBTitle: crystal structure of ph0061
68 c2vbzA_
not modelled
96.0
19
PDB header: dna-binding proteinChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
69 c2p6tH_
not modelled
95.9
21
PDB header: transcriptionChain: H: PDB Molecule: transcriptional regulator, lrp/asnc family;PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
70 c2e7xA_
not modelled
95.9
21
PDB header: transcription regulatorChain: A: PDB Molecule: 150aa long hypothetical transcriptional regulator;PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
71 c1i1gA_
not modelled
95.7
33
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator lrpa;PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
72 c3r0aB_
not modelled
95.7
19
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator;PDBTitle: possible transcriptional regulator from methanosarcina mazei go1 (gi2 21227196)
73 c2x4hA_
not modelled
95.6
29
PDB header: transcriptionChain: A: PDB Molecule: hypothetical protein sso2273;PDBTitle: crystal structure of the hypothetical protein sso2273 from2 sulfolobus solfataricus
74 c2ewnA_
not modelled
95.5
15
PDB header: ligase, transcriptionChain: A: PDB Molecule: bira bifunctional protein;PDBTitle: ecoli biotin repressor with co-repressor analog
75 c2cfxD_
not modelled
95.3
9
PDB header: transcriptionChain: D: PDB Molecule: hth-type transcriptional regulator lrpc;PDBTitle: structure of b.subtilis lrpc
76 d2oola1
not modelled
95.3
12
Fold: Profilin-likeSuperfamily: GAF domain-likeFamily: GAF domain77 c2ia0A_
not modelled
95.2
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hth-type transcriptional regulator pf0864;PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
78 c3cuoB_
not modelled
95.2
20
PDB header: transcription regulatorChain: B: PDB Molecule: uncharacterized hth-type transcriptional regulator ygav;PDBTitle: crystal structure of the predicted dna-binding transcriptional2 regulator from e. coli
79 c6paxA_
not modelled
95.0
19
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeobox protein pax-6;PDBTitle: crystal structure of the human pax-6 paired domain-dna2 complex reveals a general model for pax protein-dna3 interactions
80 c2fa5B_
not modelled
95.0
7
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator marr/emrr family;PDBTitle: the crystal structure of an unliganded multiple antibiotic-2 resistance repressor (marr) from xanthomonas campestris
81 c1b9nA_
not modelled
94.9
15
PDB header: transcriptionChain: A: PDB Molecule: protein (mode);PDBTitle: regulator from escherichia coli
82 d1r1ua_
not modelled
94.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators83 d1sfxa_
not modelled
94.9
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: TrmB-like84 d1r71a_
not modelled
94.9
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: KorB DNA-binding domain-likeFamily: KorB DNA-binding domain-like85 d1pdnc_
not modelled
94.8
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain86 c2r0qF_
not modelled
94.8
21
PDB header: recombination/dnaChain: F: PDB Molecule: putative transposon tn552 dna-invertase bin3;PDBTitle: crystal structure of a serine recombinase- dna regulatory2 complex
87 c1r71B_
not modelled
94.7
28
PDB header: transcription/dnaChain: B: PDB Molecule: transcriptional repressor protein korb;PDBTitle: crystal structure of the dna binding domain of korb in2 complex with the operator dna
88 c3tgnA_
not modelled
94.7
29
PDB header: transcriptionChain: A: PDB Molecule: adc operon repressor adcr;PDBTitle: crystal structure of the zinc-dependent marr family transcriptional2 regulator adcr in the zn(ii)-bound state
89 c3i4pA_
not modelled
94.7
19
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, asnc family;PDBTitle: crystal structure of asnc family transcriptional regulator from2 agrobacterium tumefaciens
90 c2vn2B_
not modelled
94.7
17
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
91 d1r1ta_
not modelled
94.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators92 d1lnwa_
not modelled
94.6
3
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators93 d1okra_
not modelled
94.6
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Penicillinase repressor94 d1k78a1
not modelled
94.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain95 c2it0A_
not modelled
94.5
30
PDB header: transcription/dnaChain: A: PDB Molecule: iron-dependent repressor ider;PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
96 c1r22B_
not modelled
94.4
15
PDB header: transcription repressorChain: B: PDB Molecule: transcriptional repressor smtb;PDBTitle: crystal structure of the cyanobacterial metallothionein2 repressor smtb (c14s/c61s/c121s mutant) in the zn2alpha5-3 form
97 c2gm4B_
not modelled
94.4
19
PDB header: recombination, dnaChain: B: PDB Molecule: transposon gamma-delta resolvase;PDBTitle: an activated, tetrameric gamma-delta resolvase: hin chimaera bound to2 cleaved dna
98 d1gxqa_
not modelled
94.4
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like99 c2oqgA_
not modelled
94.4
22
PDB header: transcriptionChain: A: PDB Molecule: possible transcriptional regulator, arsr family protein;PDBTitle: arsr-like transcriptional regulator from rhodococcus sp. rha1
100 c3frwF_
not modelled
94.4
19
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative trp repressor protein;PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
101 d2o9ca1
not modelled
94.3
8
Fold: Profilin-likeSuperfamily: GAF domain-likeFamily: GAF domain102 c2zxjB_
not modelled
94.3
18
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein walr;PDBTitle: crystal structure of yycf dna-binding domain from staphylococcus2 aureus
103 d1vz0a1
not modelled
94.2
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: KorB DNA-binding domain-likeFamily: KorB DNA-binding domain-like104 d6paxa1
not modelled
94.2
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain105 c2hwvA_
not modelled
94.2
23
PDB header: transcriptionChain: A: PDB Molecule: dna-binding response regulator vicr;PDBTitle: crystal structure of an essential response regulator dna binding2 domain, vicrc in enterococcus faecalis, a member of the yycf3 subfamily.
106 c3korD_
not modelled
94.0
22
PDB header: transcriptionChain: D: PDB Molecule: possible trp repressor;PDBTitle: crystal structure of a putative trp repressor from staphylococcus2 aureus
107 c2x48B_
not modelled
94.0
28
PDB header: viral proteinChain: B: PDB Molecule: cag38821;PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
108 c3trcA_
not modelled
93.9
8
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of the gaf domain from a phosphoenolpyruvate-protein2 phosphotransferase (ptsp) from coxiella burnetii
109 c2v79B_
not modelled
93.9
19
PDB header: dna-binding proteinChain: B: PDB Molecule: dna replication protein dnad;PDBTitle: crystal structure of the n-terminal domain of dnad from2 bacillus subtilis
110 c3f6oB_
not modelled
93.9
17
PDB header: transcription regulatorChain: B: PDB Molecule: probable transcriptional regulator, arsr familyPDBTitle: crystal structure of arsr family transcriptional regulator,2 rha00566
111 d2p4wa1
not modelled
93.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF1790-like112 c3nqoB_
not modelled
93.8
23
PDB header: transcriptionChain: B: PDB Molecule: marr-family transcriptional regulator;PDBTitle: crystal structure of a marr family transcriptional regulator (cd1569)2 from clostridium difficile 630 at 2.20 a resolution
113 c2elhA_
not modelled
93.8
21
PDB header: dna binding proteinChain: A: PDB Molecule: cg11849-pa;PDBTitle: solution structure of the cenp-b n-terminal dna-binding2 domain of fruit fly distal antenna cg11849-pa
114 c1f5tA_
not modelled
93.8
33
PDB header: transcription/dnaChain: A: PDB Molecule: diphtheria toxin repressor;PDBTitle: diphtheria tox repressor (c102d mutant) complexed with2 nickel and dtxr consensus binding sequence
115 d3broa1
not modelled
93.7
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators116 d2a61a1
not modelled
93.7
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators117 c2nyxB_
not modelled
93.6
11
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulatory protein, rv1404;PDBTitle: crystal structure of rv1404 from mycobacterium tuberculosis
118 d2g9wa1
not modelled
93.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Penicillinase repressor119 c2nnnB_
not modelled
93.5
8
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulator;PDBTitle: crystal structure of probable transcriptional regulator from2 pseudomonas aeruginosa
120 d1kgsa1
not modelled
93.4
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like