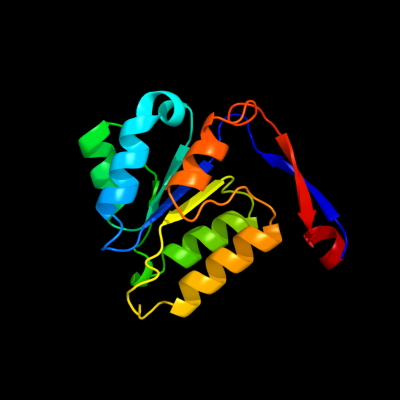
| 1 | d1b93a_
|
|
 |
100.0 |
100 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
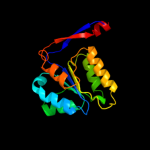
| 2 | d1vmda_
|
|
 |
100.0 |
54 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
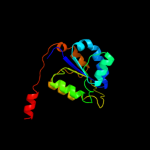
| 3 | d1wo8a1
|
|
 |
100.0 |
45 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
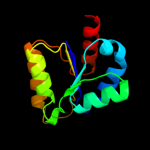
| 4 | d1a9xa2
|
|
 |
100.0 |
15 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 5 | c2yvqA_
|
|
 |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 6 | c1m6vE_
|
|
 |
99.9 |
14 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
| 7 | d1zcza1
|
|
 |
98.9 |
19 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 8 | d1pkxa1
|
|
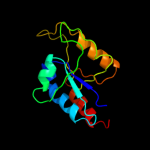 |
98.9 |
22 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 9 | d1g8ma1
|
|
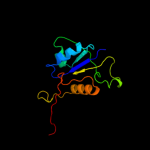 |
98.7 |
21 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 10 | c1zczA_
|
|
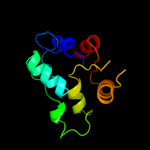 |
98.4 |
19 |
PDB header:transferase/hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of phosphoribosylaminoimidazolecarboxamide2 formyltransferase / imp cyclohydrolase (tm1249) from thermotoga3 maritima at 1.88 a resolution
|
| 11 | c4a1oB_
|
|
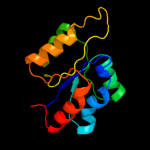 |
98.3 |
19 |
PDB header:transferase-hydrolase
Chain: B: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
|
| 12 | c1thzA_
|
|
 |
97.6 |
28 |
PDB header:transferase, hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of avian aicar transformylase in complex2 with a novel inhibitor identified by virtual ligand3 screening
|
| 13 | d1ytla1
|
|
 |
87.1 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:ACDE2-like |
| 14 | c2o48X_
|
|
 |
83.3 |
14 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 15 | c3ec7C_
|
|
 |
80.3 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase;
PDBTitle: crystal structure of putative dehydrogenase from salmonella2 typhimurium lt2
|
| 16 | d2a0ua1
|
|
 |
80.2 |
16 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 17 | d1t0kb_
|
|
 |
76.7 |
18 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 18 | c3cpqB_
|
|
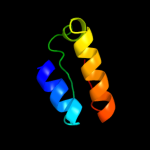 |
76.2 |
18 |
PDB header:ribosomal protein
Chain: B: PDB Molecule:50s ribosomal protein l30e;
PDBTitle: crystal structure of l30e a ribosomal protein from2 methanocaldococcus jannaschii dsm2661 (mj1044)
|
| 19 | d1w41a1
|
|
 |
75.9 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 20 | d2bo1a1
|
|
 |
72.8 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 21 | c4a1dG_ |
|
not modelled |
72.2 |
15 |
PDB header:ribosome
Chain: G: PDB Molecule:rpl30;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
|
| 22 | c2i7pA_ |
|
not modelled |
71.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:pantothenate kinase 3;
PDBTitle: crystal structure of human pank3 in complex with accoa
|
| 23 | c3q2kB_ |
|
not modelled |
70.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
| 24 | c2vpnB_ |
|
not modelled |
70.9 |
10 |
PDB header:transport
Chain: B: PDB Molecule:periplasmic substrate binding protein;
PDBTitle: high-resolution structure of the periplasmic ectoine-2 binding protein from teaabc trap-transporter of halomonas3 elongata
|
| 25 | c3ng3A_ |
|
not modelled |
69.1 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of deoxyribose phosphate aldolase from mycobacterium2 avium 104 in a schiff base with an unknown aldehyde
|
| 26 | d1t9ka_ |
|
not modelled |
68.8 |
23 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 27 | d1n7ka_ |
|
not modelled |
68.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 28 | c2q5cA_ |
|
not modelled |
68.3 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 29 | c3fxbB_ |
|
not modelled |
66.7 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:trap dicarboxylate transporter, dctp subunit;
PDBTitle: crystal structure of the ectoine-binding protein ueha
|
| 30 | c2zkr6_ |
|
not modelled |
64.4 |
20 |
PDB header:ribosomal protein/rna
Chain: 6: PDB Molecule:60s ribosomal protein l30e;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 31 | c3nklA_ |
|
not modelled |
63.2 |
12 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
| 32 | c2pfzA_ |
|
not modelled |
61.8 |
4 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of dctp6, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
|
| 33 | c2lbwA_ |
|
not modelled |
61.4 |
14 |
PDB header:rna binding protein
Chain: A: PDB Molecule:h/aca ribonucleoprotein complex subunit 2;
PDBTitle: solution structure of the s. cerevisiae h/aca rnp protein nhp2p-s82w2 mutant
|
| 34 | d1t5oa_ |
|
not modelled |
59.3 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 35 | d2ctza1 |
|
not modelled |
58.9 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 36 | c1evjC_ |
|
not modelled |
58.8 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: crystal structure of glucose-fructose oxidoreductase (gfor)2 delta1-22 s64d
|
| 37 | c2pfyA_ |
|
not modelled |
57.8 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of dctp7, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
|
| 38 | c2jlhA_ |
|
not modelled |
57.7 |
11 |
PDB header:protein transport
Chain: A: PDB Molecule:yop proteins translocation protein u;
PDBTitle: crystal structure of the cytoplasmic domain of yersinia2 pestis yscu n263a mutant
|
| 39 | c3on1A_ |
|
not modelled |
56.1 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:bh2414 protein;
PDBTitle: the structure of a protein with unknown function from bacillus2 halodurans c
|
| 40 | c2yvkA_ |
|
not modelled |
56.0 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
| 41 | c1zh8B_ |
|
not modelled |
55.7 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 42 | c3r6uA_ |
|
not modelled |
55.7 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:choline-binding protein;
PDBTitle: crystal structure of choline binding protein opubc from bacillus2 subtilis
|
| 43 | c3pppA_ |
|
not modelled |
55.5 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine/carnitine/choline-binding protein;
PDBTitle: structures of the substrate-binding protein provide insights into the2 multiple compatible solutes binding specificities of bacillus3 subtilis abc transporter opuc
|
| 44 | c3t7yB_ |
|
not modelled |
53.1 |
16 |
PDB header:protein transport
Chain: B: PDB Molecule:yop proteins translocation protein u;
PDBTitle: structure of an autocleavage-inactive mutant of the cytoplasmic domain2 of ct091, the yscu homologue of chlamydia trachomatis
|
| 45 | c3o3nB_ |
|
not modelled |
52.9 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:beta-subunit 2-hydroxyacyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
| 46 | c3btuD_ |
|
not modelled |
52.5 |
15 |
PDB header:transcription
Chain: D: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal structure of the super-repressor mutant of gal80p2 from saccharomyces cerevisiae; gal80(s2) [e351k]
|
| 47 | c1ofgF_ |
|
not modelled |
50.9 |
16 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 48 | c1h6dL_ |
|
not modelled |
50.9 |
16 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 49 | c2cb1A_ |
|
not modelled |
49.8 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:o-acetyl homoserine sulfhydrylase;
PDBTitle: crystal structure of o-actetyl homoserine sulfhydrylase2 from thermus thermophilus hb8,oah2.
|
| 50 | c2f5xC_ |
|
not modelled |
49.8 |
19 |
PDB header:transport protein
Chain: C: PDB Molecule:bugd;
PDBTitle: structure of periplasmic binding protein bugd
|
| 51 | c3v5nA_ |
|
not modelled |
49.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of oxidoreductase from sinorhizobium meliloti
|
| 52 | d3bzra1 |
|
not modelled |
48.8 |
18 |
Fold:EscU C-terminal domain-like
Superfamily:EscU C-terminal domain-like
Family:EscU C-terminal domain-like |
| 53 | c3bzrA_ |
|
not modelled |
48.8 |
18 |
PDB header:membrane protein, protein transport
Chain: A: PDB Molecule:escu;
PDBTitle: crystal structure of escu c-terminal domain with n262d mutation, space2 group p 41 21 2
|
| 54 | c3c01H_ |
|
not modelled |
48.3 |
16 |
PDB header:membrane protein, protein transport
Chain: H: PDB Molecule:surface presentation of antigens protein spas;
PDBTitle: crystal structural of native spas c-terminal domain
|
| 55 | c2zzxD_ |
|
not modelled |
48.3 |
17 |
PDB header:transport protein
Chain: D: PDB Molecule:abc transporter, solute-binding protein;
PDBTitle: crystal structure of a periplasmic substrate binding protein in2 complex with lactate
|
| 56 | d2nvwa1 |
|
not modelled |
47.6 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 57 | c2vt1B_ |
|
not modelled |
47.6 |
18 |
PDB header:membrane protein
Chain: B: PDB Molecule:surface presentation of antigens protein spas;
PDBTitle: crystal structure of the cytoplasmic domain of spa40, the2 specificity switch for the shigella flexneri type iii3 secretion system
|
| 58 | c3c00B_ |
|
not modelled |
47.3 |
18 |
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:escu;
PDBTitle: crystal structural of the mutated g247t escu/spas c-terminal domain
|
| 59 | d1rlga_ |
|
not modelled |
45.7 |
18 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 60 | c2xznU_ |
|
not modelled |
45.2 |
12 |
PDB header:ribosome
Chain: U: PDB Molecule:ribosomal protein l7ae containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 61 | d1jx7a_ |
|
not modelled |
44.3 |
20 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 62 | c3b50A_ |
|
not modelled |
43.9 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:sialic acid-binding periplasmic protein siap;
PDBTitle: structure of h. influenzae sialic acid binding protein2 bound to neu5ac.
|
| 63 | c3o3nA_ |
|
not modelled |
43.7 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:alpha-subunit 2-hydroxyisocaproyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
| 64 | c2hpgB_ |
|
not modelled |
41.9 |
15 |
PDB header:ligand binding protein
Chain: B: PDB Molecule:abc transporter, periplasmic substrate-binding
PDBTitle: the crystal structure of a thermophilic trap periplasmic2 binding protein
|
| 65 | d1su7a_ |
|
not modelled |
41.9 |
16 |
Fold:Prismane protein-like
Superfamily:Prismane protein-like
Family:Carbon monoxide dehydrogenase |
| 66 | c2hzkB_ |
|
not modelled |
39.1 |
20 |
PDB header:ligand binding, transport protein
Chain: B: PDB Molecule:trap-t family sorbitol/mannitol transporter, periplasmic
PDBTitle: crystal structures of a sodium-alpha-keto acid binding subunit from a2 trap transporter in its open form
|
| 67 | c1toaA_ |
|
not modelled |
37.7 |
22 |
PDB header:binding protein
Chain: A: PDB Molecule:protein (periplasmic binding protein troa);
PDBTitle: periplasmic zinc binding protein troa from treponema pallidum
|
| 68 | d1toaa_ |
|
not modelled |
37.7 |
22 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
| 69 | c1ibjC_ |
|
not modelled |
37.2 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:cystathionine beta-lyase;
PDBTitle: crystal structure of cystathionine beta-lyase from arabidopsis2 thaliana
|
| 70 | d1ibja_ |
|
not modelled |
37.2 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 71 | c3e6gA_ |
|
not modelled |
36.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:cystathionine gamma-lyase-like protein;
PDBTitle: crystal structure of xometc, a cystathionine c-lyase-like2 protein from xanthomonas oryzae pv.oryzae
|
| 72 | c2dvzA_ |
|
not modelled |
36.7 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: structure of a periplasmic transporter
|
| 73 | c3g13B_ |
|
not modelled |
36.4 |
22 |
PDB header:recombination
Chain: B: PDB Molecule:putative conjugative transposon recombinase;
PDBTitle: crystal structure of putative conjugative transposon recombinase from2 clostridium difficile
|
| 74 | c2pjuD_ |
|
not modelled |
36.2 |
9 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 75 | d1rxwa2 |
|
not modelled |
35.5 |
19 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 76 | c3aemD_ |
|
not modelled |
35.4 |
12 |
PDB header:lyase
Chain: D: PDB Molecule:methionine gamma-lyase;
PDBTitle: reaction intermediate structure of entamoeba histolytica methionine2 gamma-lyase 1 containing michaelis complex and methionine imine-3 pyridoxamine-5'-phosphate
|
| 77 | d1y4ia1 |
|
not modelled |
34.8 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 78 | c3ce8A_ |
|
not modelled |
34.5 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:putative pii-like nitrogen regulatory protein;
PDBTitle: crystal structure of a duf3240 family protein (sbal_0098) from2 shewanella baltica os155 at 2.40 a resolution
|
| 79 | c3ngjC_ |
|
not modelled |
33.4 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 entamoeba histolytica
|
| 80 | d1vqof1 |
|
not modelled |
32.9 |
20 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 81 | d2aifa1 |
|
not modelled |
32.7 |
14 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 82 | c3ndnC_ |
|
not modelled |
32.1 |
27 |
PDB header:lyase
Chain: C: PDB Molecule:o-succinylhomoserine sulfhydrylase;
PDBTitle: crystal structure of o-succinylhomoserine sulfhydrylase from2 mycobacterium tuberculosis covalently bound to pyridoxal-5-phosphate
|
| 83 | d1v77a_ |
|
not modelled |
31.7 |
18 |
Fold:7-stranded beta/alpha barrel
Superfamily:PHP domain-like
Family:RNase P subunit p30 |
| 84 | c3mc6C_ |
|
not modelled |
31.6 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of scdpl1
|
| 85 | c3o85A_ |
|
not modelled |
31.3 |
16 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:ribosomal protein l7ae;
PDBTitle: giardia lamblia 15.5kd rna binding protein
|
| 86 | c3moiA_ |
|
not modelled |
29.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
| 87 | d2czwa1 |
|
not modelled |
29.6 |
24 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 88 | d1gc0a_ |
|
not modelled |
29.2 |
24 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 89 | d1qgna_ |
|
not modelled |
29.1 |
21 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 90 | d1mc8a2 |
|
not modelled |
28.9 |
21 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 91 | d2bwna1 |
|
not modelled |
28.8 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 92 | c3jpyA_ |
|
not modelled |
27.9 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:glutamate [nmda] receptor subunit epsilon-2;
PDBTitle: crystal structure of the zinc-bound amino terminal domain of the nmda2 receptor subunit nr2b
|
| 93 | d1ul1x2 |
|
not modelled |
27.7 |
27 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 94 | c3h14A_ |
|
not modelled |
27.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase, classes i and ii;
PDBTitle: crystal structure of a putative aminotransferase from silicibacter2 pomeroyi
|
| 95 | c3ezsB_ |
|
not modelled |
27.3 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase aspb;
PDBTitle: crystal structure of aminotransferase aspb (np_207418.1) from2 helicobacter pylori 26695 at 2.19 a resolution
|
| 96 | d2ozba1 |
|
not modelled |
26.9 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 97 | c2nvwB_ |
|
not modelled |
25.7 |
15 |
PDB header:transcription
Chain: B: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
|
| 98 | d1p1xa_ |
|
not modelled |
25.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 99 | d1fc4a_ |
|
not modelled |
25.4 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 100 | d1b43a2 |
|
not modelled |
25.2 |
16 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 101 | c1o94D_ |
|
not modelled |
24.9 |
14 |
PDB header:electron transport
Chain: D: PDB Molecule:electron transfer flavoprotein alpha-subunit;
PDBTitle: ternary complex between trimethylamine dehydrogenase and2 electron transferring flavoprotein
|
| 102 | d2fc3a1 |
|
not modelled |
24.8 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 103 | d1ydwa1 |
|
not modelled |
24.1 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 104 | c3h5oB_ |
|
not modelled |
24.1 |
14 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator gntr;
PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
|
| 105 | c3ftqA_ |
|
not modelled |
24.0 |
18 |
PDB header:cell cycle
Chain: A: PDB Molecule:septin-2;
PDBTitle: crystal structure of septin 2 in complex with gppnhp and2 mg2+
|
| 106 | c2hdyA_ |
|
not modelled |
23.7 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:selenocysteine lyase;
PDBTitle: structure of human selenocysteine lyase
|
| 107 | c3gyyC_ |
|
not modelled |
23.4 |
10 |
PDB header:transport protein
Chain: C: PDB Molecule:periplasmic substrate binding protein;
PDBTitle: the ectoine binding protein of the teaabc trap transporter teaa in the2 apo-state
|
| 108 | d1a77a2 |
|
not modelled |
23.2 |
16 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 109 | c1yl7F_ |
|
not modelled |
23.1 |
25 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: the crystal structure of mycobacterium tuberculosis2 dihydrodipicolinate reductase (rv2773c) in complex with nadh (crystal3 form c)
|
| 110 | c2zkrf_ |
|
not modelled |
23.0 |
26 |
PDB header:ribosomal protein/rna
Chain: F: PDB Molecule:rna expansion segment es7 part iii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 111 | c3nysA_ |
|
not modelled |
22.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase wbpe;
PDBTitle: x-ray structure of the k185a mutant of wbpe (wlbe) from pseudomonas2 aeruginosa in complex with plp at 1.45 angstrom resolution
|
| 112 | c1a77A_ |
|
not modelled |
22.1 |
21 |
PDB header:5'-3' exo/endo nuclease
Chain: A: PDB Molecule:flap endonuclease-1 protein;
PDBTitle: flap endonuclease-1 from methanococcus jannaschii
|
| 113 | d2alea1 |
|
not modelled |
22.1 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 114 | c3mfqB_ |
|
not modelled |
21.9 |
15 |
PDB header:metal binding protein
Chain: B: PDB Molecule:high-affinity zinc uptake system protein znua;
PDBTitle: a glance into the metal binding specificity of troa: where elaborate2 behaviors occur in the active center
|
| 115 | c3ceaA_ |
|
not modelled |
21.7 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 116 | c3g0tA_ |
|
not modelled |
21.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative aminotransferase;
PDBTitle: crystal structure of putative aspartate aminotransferase (np_905498.1)2 from porphyromonas gingivalis w83 at 1.75 a resolution
|
| 117 | c3f9tB_ |
|
not modelled |
21.0 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:l-tyrosine decarboxylase mfna;
PDBTitle: crystal structure of l-tyrosine decarboxylase mfna (ec 4.1.1.25)2 (np_247014.1) from methanococcus jannaschii at 2.11 a resolution
|
| 118 | c1b43A_ |
|
not modelled |
20.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:protein (fen-1);
PDBTitle: fen-1 from p. furiosus
|
| 119 | c3o66A_ |
|
not modelled |
20.4 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine/carnitine/choline abc transporter;
PDBTitle: crystal structure of glycine betaine/carnitine/choline abc transporter
|
| 120 | c2p0oA_ |
|
not modelled |
20.2 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein duf871;
PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
|