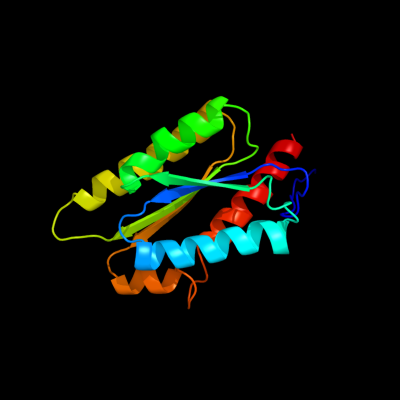
| 1 | c3mk3L_
|
|
 |
100.0 |
91 |
PDB header:transferase
Chain: L: PDB Molecule:6,7-dimethyl-8-ribityllumazine synthase;
PDBTitle: crystal structure of lumazine synthase from salmonella typhimurium lt2
|
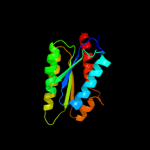
| 2 | d1rvv1_
|
|
 |
100.0 |
53 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
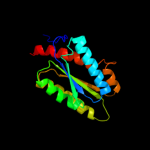
| 3 | d1nqua_
|
|
 |
100.0 |
50 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
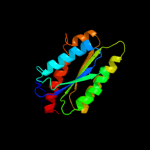
| 4 | d1c2ya_
|
|
 |
100.0 |
50 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
| 5 | d1c41a_
|
|
 |
100.0 |
43 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
| 6 | d1kz1a_
|
|
 |
100.0 |
38 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
| 7 | d1ejba_
|
|
 |
100.0 |
35 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
| 8 | d1di0a_
|
|
 |
100.0 |
32 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
| 9 | c2f59B_
|
|
 |
100.0 |
40 |
PDB header:transferase
Chain: B: PDB Molecule:6,7-dimethyl-8-ribityllumazine synthase 1;
PDBTitle: lumazine synthase ribh1 from brucella abortus (gene bruab1_0785,2 swiss-prot entry q57dy1) complexed with inhibitor 5-nitro-6-(d-3 ribitylamino)-2,4(1h,3h) pyrimidinedione
|
| 10 | c2obxH_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: H: PDB Molecule:6,7-dimethyl-8-ribityllumazine synthase 1;
PDBTitle: lumazine synthase ribh2 from mesorhizobium loti (gene mll7281, swiss-2 prot entry q986n2) complexed with inhibitor 5-nitro-6-(d-3 ribitylamino)-2,4(1h,3h) pyrimidinedione
|
| 11 | c1w19E_
|
|
 |
100.0 |
45 |
PDB header:transferase
Chain: E: PDB Molecule:6,7-dimethyl-8-ribityllumazine synthase;
PDBTitle: lumazine synthase from mycobacterium tuberculosis bound to2 3-(1,3,7-trihydro-9-d-ribityl-2,6,8-purinetrione-7-yl)3 propane 1-phosphate
|
| 12 | c2b99A_
|
|
 |
97.7 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase;
PDBTitle: crystal structure of an archaeal pentameric riboflavin2 synthase complex with a substrate analog inhibitor
|
| 13 | c3lftA_
|
|
 |
94.9 |
16 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the abc domain in complex with l-trp from2 streptococcus pneumonia to 1.35a
|
| 14 | c2qh8A_
|
|
 |
92.2 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of conserved domain protein from vibrio2 cholerae o1 biovar eltor str. n16961
|
| 15 | c3h5oB_
|
|
 |
90.9 |
17 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator gntr;
PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
|
| 16 | c3kkeA_
|
|
 |
90.7 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:laci family transcriptional regulator;
PDBTitle: crystal structure of a laci family transcriptional regulator2 from mycobacterium smegmatis
|
| 17 | d1guda_
|
|
 |
90.6 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 18 | d1gcaa_
|
|
 |
89.3 |
13 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 19 | c3ksmA_
|
|
 |
88.1 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
| 20 | c3ma0A_
|
|
 |
87.8 |
9 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:d-xylose-binding periplasmic protein;
PDBTitle: closed liganded crystal structure of xylose binding protein from2 escherichia coli
|
| 21 | c3bilA_ |
|
not modelled |
87.6 |
6 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable laci-family transcriptional regulator;
PDBTitle: crystal structure of a probable laci family transcriptional2 regulator from corynebacterium glutamicum
|
| 22 | c3l6uA_ |
|
not modelled |
86.6 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system periplasmic
PDBTitle: crystal structure of abc-type sugar transport system,2 periplasmic component from exiguobacterium sibiricum
|
| 23 | c3o74A_ |
|
not modelled |
86.1 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:fructose transport system repressor frur;
PDBTitle: crystal structure of cra transcriptional dual regulator from2 pseudomonas putida
|
| 24 | c2fn9A_ |
|
not modelled |
86.0 |
24 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:ribose abc transporter, periplasmic ribose-binding protein;
PDBTitle: thermotoga maritima ribose binding protein unliganded form
|
| 25 | c3hcwB_ |
|
not modelled |
85.8 |
13 |
PDB header:rna binding protein
Chain: B: PDB Molecule:maltose operon transcriptional repressor;
PDBTitle: crystal structure of probable maltose operon transcriptional repressor2 malr from staphylococcus areus
|
| 26 | c3o1hB_ |
|
not modelled |
84.8 |
17 |
PDB header:signaling protein
Chain: B: PDB Molecule:periplasmic protein tort;
PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
|
| 27 | d8abpa_ |
|
not modelled |
83.8 |
16 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 28 | c3brqA_ |
|
not modelled |
82.8 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator ascg;
PDBTitle: crystal structure of the escherichia coli transcriptional repressor2 ascg
|
| 29 | c3e3mA_ |
|
not modelled |
82.6 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of a laci family transcriptional2 regulator from silicibacter pomeroyi
|
| 30 | c3mizB_ |
|
not modelled |
82.4 |
14 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator protein, laci
PDBTitle: crystal structure of a putative transcriptional regulator2 protein, lacl family from rhizobium etli
|
| 31 | c3c3kA_ |
|
not modelled |
82.2 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of an uncharacterized protein from actinobacillus2 succinogenes
|
| 32 | c3ctpB_ |
|
not modelled |
81.9 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of periplasmic binding protein/laci transcriptional2 regulator from alkaliphilus metalliredigens qymf complexed with d-3 xylulofuranose
|
| 33 | c2qu7B_ |
|
not modelled |
81.6 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of a putative transcription regulator2 from staphylococcus saprophyticus subsp. saprophyticus
|
| 34 | d2dria_ |
|
not modelled |
80.9 |
19 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 35 | c2vk2A_ |
|
not modelled |
80.7 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter periplasmic-binding protein ytfq;
PDBTitle: crystal structure of a galactofuranose binding protein
|
| 36 | c3d8uA_ |
|
not modelled |
79.3 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:purr transcriptional regulator;
PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
|
| 37 | d2fvya1 |
|
not modelled |
76.6 |
11 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 38 | d1byka_ |
|
not modelled |
76.4 |
18 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 39 | d1dbqa_ |
|
not modelled |
75.6 |
8 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 40 | d1y5ea1 |
|
not modelled |
74.8 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 41 | c3gv0A_ |
|
not modelled |
74.1 |
17 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
|
| 42 | c2o20H_ |
|
not modelled |
71.7 |
11 |
PDB header:transcription
Chain: H: PDB Molecule:catabolite control protein a;
PDBTitle: crystal structure of transcription regulator ccpa of lactococcus2 lactis
|
| 43 | d1tjya_ |
|
not modelled |
71.5 |
24 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 44 | c2rjoA_ |
|
not modelled |
71.0 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:twin-arginine translocation pathway signal protein;
PDBTitle: crystal structure of twin-arginine translocation pathway signal2 protein from burkholderia phytofirmans
|
| 45 | c3egcF_ |
|
not modelled |
70.6 |
11 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative ribose operon repressor;
PDBTitle: crystal structure of a putative ribose operon repressor from2 burkholderia thailandensis
|
| 46 | c3k9cA_ |
|
not modelled |
70.1 |
20 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family protein;
PDBTitle: crystal structure of laci transcriptional regulator from rhodococcus2 species.
|
| 47 | c3l49D_ |
|
not modelled |
69.7 |
13 |
PDB header:transport protein
Chain: D: PDB Molecule:abc sugar (ribose) transporter, periplasmic
PDBTitle: crystal structure of abc sugar transporter subunit from2 rhodobacter sphaeroides 2.4.1
|
| 48 | d2nzug1 |
|
not modelled |
68.6 |
18 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 49 | c3clkB_ |
|
not modelled |
67.8 |
17 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcription regulator;
PDBTitle: crystal structure of a transcription regulator from lactobacillus2 plantarum
|
| 50 | c3jy6B_ |
|
not modelled |
67.8 |
15 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci transcriptional regulator from lactobacillus2 brevis
|
| 51 | c2h0aA_ |
|
not modelled |
67.4 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of probable transcription regulator from2 thermus thermophilus
|
| 52 | c2iksA_ |
|
not modelled |
66.5 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:dna-binding transcriptional dual regulator;
PDBTitle: crystal structure of n-terminal truncated dna-binding transcriptional2 dual regulator from escherichia coli k12
|
| 53 | d1uuya_ |
|
not modelled |
65.8 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 54 | c3e61A_ |
|
not modelled |
65.0 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative transcriptional repressor of ribose operon;
PDBTitle: crystal structure of a putative transcriptional repressor of ribose2 operon from staphylococcus saprophyticus subsp. saprophyticus
|
| 55 | c3bblA_ |
|
not modelled |
64.9 |
11 |
PDB header:regulatory protein
Chain: A: PDB Molecule:regulatory protein of laci family;
PDBTitle: crystal structure of a regulatory protein of laci family from2 chloroflexus aggregans
|
| 56 | c2fqxA_ |
|
not modelled |
64.5 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:membrane lipoprotein tmpc;
PDBTitle: pnra from treponema pallidum complexed with guanosine
|
| 57 | c3rotA_ |
|
not modelled |
63.2 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:abc sugar transporter, periplasmic sugar binding protein;
PDBTitle: crystal structure of abc sugar transporter (periplasmic sugar binding2 protein) from legionella pneumophila
|
| 58 | d2f7wa1 |
|
not modelled |
62.5 |
14 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 59 | c3cs3A_ |
|
not modelled |
61.4 |
18 |
PDB header:transcription regulator
Chain: A: PDB Molecule:sugar-binding transcriptional regulator, laci family;
PDBTitle: crystal structure of sugar-binding transcriptional regulator (laci2 family) from enterococcus faecalis
|
| 60 | c2ioyB_ |
|
not modelled |
61.3 |
17 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:periplasmic sugar-binding protein;
PDBTitle: crystal structure of thermoanaerobacter tengcongensis2 ribose binding protein
|
| 61 | d1tlfa_ |
|
not modelled |
59.9 |
11 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 62 | d1jyea_ |
|
not modelled |
59.1 |
10 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 63 | c1jyeA_ |
|
not modelled |
59.1 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:lactose operon repressor;
PDBTitle: structure of a dimeric lac repressor with c-terminal deletion and k84l2 substitution
|
| 64 | c3huuC_ |
|
not modelled |
58.7 |
14 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcription regulator like protein;
PDBTitle: crystal structure of transcription regulator like protein from2 staphylococcus haemolyticus
|
| 65 | c2x7xA_ |
|
not modelled |
58.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: fructose binding periplasmic domain of hybrid two component2 system bt1754
|
| 66 | c3qk7C_ |
|
not modelled |
56.9 |
18 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcriptional regulator from yersinia2 pestis biovar microtus str. 91001
|
| 67 | d1jlja_ |
|
not modelled |
55.2 |
14 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 68 | c3lq1A_ |
|
not modelled |
53.1 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-
PDBTitle: crystal structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene2 1-carboxylic acid synthase/2-oxoglutarate decarboxylase3 from listeria monocytogenes str. 4b f2365
|
| 69 | c3k4hA_ |
|
not modelled |
48.6 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator laci from2 bacillus cereus subsp. cytotoxis nvh 391-98
|
| 70 | c2is8A_ |
|
not modelled |
48.2 |
18 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
| 71 | d2csua3 |
|
not modelled |
48.1 |
20 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 72 | c2w7tA_ |
|
not modelled |
46.4 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:putative cytidine triphosphate synthase;
PDBTitle: trypanosoma brucei ctps - glutaminase domain with bound2 acivicin
|
| 73 | c2csuB_ |
|
not modelled |
46.3 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
| 74 | c3g1wB_ |
|
not modelled |
45.9 |
13 |
PDB header:transport protein
Chain: B: PDB Molecule:sugar abc transporter;
PDBTitle: crystal structure of sugar abc transporter (sugar-binding protein)2 from bacillus halodurans
|
| 75 | d1di6a_ |
|
not modelled |
45.6 |
11 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 76 | c2nu9E_ |
|
not modelled |
45.1 |
21 |
PDB header:ligase
Chain: E: PDB Molecule:succinyl-coa synthetase beta chain;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase2 orthorhombic crystal form
|
| 77 | c2jlaD_ |
|
not modelled |
43.2 |
33 |
PDB header:transferase
Chain: D: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: crystal structure of e.coli mend, 2-succinyl-5-enolpyruvyl-2 6-hydroxy-3-cyclohexadiene-1-carboxylate synthase - semet3 protein
|
| 78 | d2nu7b1 |
|
not modelled |
42.8 |
21 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 79 | c3gybB_ |
|
not modelled |
42.4 |
17 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulators (laci-family
PDBTitle: crystal structure of a laci-family transcriptional2 regulatory protein from corynebacterium glutamicum
|
| 80 | d2a5la1 |
|
not modelled |
42.3 |
11 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 81 | c2qvcC_ |
|
not modelled |
42.2 |
12 |
PDB header:transport protein
Chain: C: PDB Molecule:sugar abc transporter, periplasmic sugar-binding
PDBTitle: crystal structure of a periplasmic sugar abc transporter2 from thermotoga maritima
|
| 82 | c2dzaA_ |
|
not modelled |
42.0 |
41 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 83 | c3c5yD_ |
|
not modelled |
41.6 |
24 |
PDB header:isomerase
Chain: D: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
|
| 84 | d1mkza_ |
|
not modelled |
41.5 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 85 | c3dbiA_ |
|
not modelled |
41.3 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:sugar-binding transcriptional regulator, laci family;
PDBTitle: crystal structure of sugar-binding transcriptional regulator (laci2 family) from escherichia coli complexed with phosphate
|
| 86 | c2rgyA_ |
|
not modelled |
38.6 |
17 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of transcriptional regulator of laci family from2 burkhoderia phymatum
|
| 87 | c3g85A_ |
|
not modelled |
38.2 |
16 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator (laci family);
PDBTitle: crystal structure of laci family transcription regulator from2 clostridium acetobutylicum
|
| 88 | d2nqra3 |
|
not modelled |
36.6 |
24 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 89 | d1u11a_ |
|
not modelled |
36.3 |
16 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 90 | d1o2da_ |
|
not modelled |
36.1 |
15 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 91 | d1b43a2 |
|
not modelled |
35.2 |
28 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 92 | d1rxwa2 |
|
not modelled |
34.4 |
22 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 93 | d2pjua1 |
|
not modelled |
34.0 |
19 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 94 | c2fw9A_ |
|
not modelled |
32.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
|
| 95 | c3jvdA_ |
|
not modelled |
32.8 |
15 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcription regulation repressor (laci2 family) from corynebacterium glutamicum
|
| 96 | d7reqa2 |
|
not modelled |
31.8 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 97 | d4pgaa_ |
|
not modelled |
31.7 |
32 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 98 | c3lp6D_ |
|
not modelled |
31.4 |
21 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
| 99 | c3kklA_ |
|
not modelled |
31.2 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable chaperone protein hsp33;
PDBTitle: crystal structure of functionally unknown hsp33 from2 saccharomyces cerevisiae
|
| 100 | d1vlja_ |
|
not modelled |
31.1 |
28 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 101 | c3brsA_ |
|
not modelled |
30.8 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of sugar transporter from clostridium2 phytofermentans
|
| 102 | c3orsD_ |
|
not modelled |
30.1 |
15 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
| 103 | c2y5sA_ |
|
not modelled |
29.8 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 104 | c3s99A_ |
|
not modelled |
29.6 |
8 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:basic membrane lipoprotein;
PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
|
| 105 | c3oryA_ |
|
not modelled |
28.8 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:flap endonuclease 1;
PDBTitle: crystal structure of flap endonuclease 1 from hyperthermophilic2 archaeon desulfurococcus amylolyticus
|
| 106 | d2jgra1 |
|
not modelled |
28.3 |
25 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
| 107 | c2ywxA_ |
|
not modelled |
27.9 |
29 |
PDB header:lyase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
|
| 108 | c1b43A_ |
|
not modelled |
27.9 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:protein (fen-1);
PDBTitle: fen-1 from p. furiosus
|
| 109 | c2hmcA_ |
|
not modelled |
27.6 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
| 110 | d1ofux_ |
|
not modelled |
27.4 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Bacterial cell division inhibitor SulA |
| 111 | c2x7jA_ |
|
not modelled |
26.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: structure of the menaquinone biosynthesis protein mend from2 bacillus subtilis
|
| 112 | d1jx6a_ |
|
not modelled |
25.3 |
12 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 113 | c2r94B_ |
|
not modelled |
25.2 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 114 | c1dl5A_ |
|
not modelled |
24.9 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:protein-l-isoaspartate o-methyltransferase;
PDBTitle: protein-l-isoaspartate o-methyltransferase
|
| 115 | c2zkiH_ |
|
not modelled |
24.0 |
8 |
PDB header:transcription
Chain: H: PDB Molecule:199aa long hypothetical trp repressor binding
PDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
|
| 116 | c1oftC_ |
|
not modelled |
23.9 |
17 |
PDB header:bacterial cell division inhibitor
Chain: C: PDB Molecule:hypothetical protein pa3008;
PDBTitle: crystal structure of sula from pseudomonas aeruginosa
|
| 117 | c3hs3A_ |
|
not modelled |
23.5 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:ribose operon repressor;
PDBTitle: crystal structure of periplasmic binding ribose operon2 repressor protein from lactobacillus acidophilus
|
| 118 | d1o7ja_ |
|
not modelled |
23.4 |
32 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 119 | d1a77a2 |
|
not modelled |
22.6 |
29 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 120 | c3okfA_ |
|
not modelled |
22.5 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: 2.5 angstrom resolution crystal structure of 3-dehydroquinate synthase2 (arob) from vibrio cholerae
|